Using ModelBuilder class for deploying PyMC models#
Motivation#
Many users face difficulty in deploying their PyMC models to production because deploying/saving/loading a user-created model is not well standardized. One of the reasons behind this is there is no direct way to save or load a model in PyMC like scikit-learn or TensorFlow. The new ModelBuilder class is aimed to improve this workflow by providing a scikit-learn inspired API to wrap your PyMC models.
The new ModelBuilder class allows users to use methods to fit(), predict(), save(), load(). Users can create any model they want, inherit the ModelBuilder class, and use predefined methods.
Let’s go through the full workflow, starting with a simple linear regression PyMC model as it’s usually written. Of course, this model is just a place-holder for your own model.
from typing import Dict, List, Optional, Tuple, Union
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc as pm
import xarray as xr
from numpy.random import RandomState
%config InlineBackend.figure_format = 'retina'
RANDOM_SEED = 8927
rng = np.random.default_rng(RANDOM_SEED)
az.style.use("arviz-darkgrid")
WARNING (pytensor.tensor.blas): Using NumPy C-API based implementation for BLAS functions.
# Generate data
x = np.linspace(start=0, stop=1, num=100)
y = 0.3 * x + 0.5 + rng.normal(0, 1, len(x))
Standard syntax#
Usually a PyMC model will have this form:
with pm.Model() as model:
# priors
a = pm.Normal("a", mu=0, sigma=1)
b = pm.Normal("b", mu=0, sigma=1)
eps = pm.HalfNormal("eps", 1.0)
# observed data
y_model = pm.Normal("y_model", mu=a + b * x, sigma=eps, observed=y)
# Fitting
idata = pm.sample()
idata.extend(pm.sample_prior_predictive())
# posterior predict
idata.extend(pm.sample_posterior_predictive(idata))
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [a, b, eps]
Sampling 2 chains for 1_000 tune and 1_000 draw iterations (2_000 + 2_000 draws total) took 4 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
Sampling: [a, b, eps, y_model]
Sampling: [y_model]
How would we deploy this model? Save the fitted model, load it on an instance, and predict? Not so simple.
ModelBuilder is built for this purpose. It is currently part of the pymc-experimental package which we can pip install with pip install pymc-experimental. As the name implies, this feature is still experimental and subject to change.
Model builder class#
Let’s import the ModelBuilder class.
from pymc_experimental.model_builder import ModelBuilder
To define our desired model we inherit from the ModelBuilder class. There are a couple of methods we need to define.
class LinearModel(ModelBuilder):
# Give the model a name
_model_type = "LinearModel"
# And a version
version = "0.1"
def build_model(self, X: pd.DataFrame, y: pd.Series, **kwargs):
"""
build_model creates the PyMC model
Parameters:
model_config: dictionary
it is a dictionary with all the parameters that we need in our model example: a_loc, a_scale, b_loc
X : pd.DataFrame
The input data that is going to be used in the model. This should be a DataFrame
containing the features (predictors) for the model. For efficiency reasons, it should
only contain the necessary data columns, not the entire available dataset, as this
will be encoded into the data used to recreate the model.
y : pd.Series
The target data for the model. This should be a Series representing the output
or dependent variable for the model.
kwargs : dict
Additional keyword arguments that may be used for model configuration.
"""
# Check the type of X and y and adjust access accordingly
X_values = X["input"].values
y_values = y.values if isinstance(y, pd.Series) else y
self._generate_and_preprocess_model_data(X_values, y_values)
with pm.Model(coords=self.model_coords) as self.model:
# Create mutable data containers
x_data = pm.MutableData("x_data", X_values)
y_data = pm.MutableData("y_data", y_values)
# prior parameters
a_mu_prior = self.model_config.get("a_mu_prior", 0.0)
a_sigma_prior = self.model_config.get("a_sigma_prior", 1.0)
b_mu_prior = self.model_config.get("b_mu_prior", 0.0)
b_sigma_prior = self.model_config.get("b_sigma_prior", 1.0)
eps_prior = self.model_config.get("eps_prior", 1.0)
# priors
a = pm.Normal("a", mu=a_mu_prior, sigma=a_sigma_prior)
b = pm.Normal("b", mu=b_mu_prior, sigma=b_sigma_prior)
eps = pm.HalfNormal("eps", eps_prior)
obs = pm.Normal("y", mu=a + b * x_data, sigma=eps, shape=x_data.shape, observed=y_data)
def _data_setter(
self, X: Union[pd.DataFrame, np.ndarray], y: Union[pd.Series, np.ndarray] = None
):
if isinstance(X, pd.DataFrame):
x_values = X["input"].values
else:
# Assuming "input" is the first column
x_values = X[:, 0]
with self.model:
pm.set_data({"x_data": x_values})
if y is not None:
pm.set_data({"y_data": y.values if isinstance(y, pd.Series) else y})
@staticmethod
def get_default_model_config() -> Dict:
"""
Returns a class default config dict for model builder if no model_config is provided on class initialization.
The model config dict is generally used to specify the prior values we want to build the model with.
It supports more complex data structures like lists, dictionaries, etc.
It will be passed to the class instance on initialization, in case the user doesn't provide any model_config of their own.
"""
model_config: Dict = {
"a_mu_prior": 0.0,
"a_sigma_prior": 1.0,
"b_mu_prior": 0.0,
"b_sigma_prior": 1.0,
"eps_prior": 1.0,
}
return model_config
@staticmethod
def get_default_sampler_config() -> Dict:
"""
Returns a class default sampler dict for model builder if no sampler_config is provided on class initialization.
The sampler config dict is used to send parameters to the sampler .
It will be used during fitting in case the user doesn't provide any sampler_config of their own.
"""
sampler_config: Dict = {
"draws": 1_000,
"tune": 1_000,
"chains": 3,
"target_accept": 0.95,
}
return sampler_config
@property
def output_var(self):
return "y"
@property
def _serializable_model_config(self) -> Dict[str, Union[int, float, Dict]]:
"""
_serializable_model_config is a property that returns a dictionary with all the model parameters that we want to save.
as some of the data structures are not json serializable, we need to convert them to json serializable objects.
Some models will need them, others can just define them to return the model_config.
"""
return self.model_config
def _save_input_params(self, idata) -> None:
"""
Saves any additional model parameters (other than the dataset) to the idata object.
These parameters are stored within `idata.attrs` using keys that correspond to the parameter names.
If you don't need to store any extra parameters, you can leave this method unimplemented.
Example:
For saving customer IDs provided as an 'customer_ids' input to the model:
self.customer_ids = customer_ids.values #this line is done outside of the function, preferably at the initialization of the model object.
idata.attrs["customer_ids"] = json.dumps(self.customer_ids.tolist()) # Convert numpy array to a JSON-serializable list.
"""
pass
pass
def _generate_and_preprocess_model_data(
self, X: Union[pd.DataFrame, pd.Series], y: Union[pd.Series, np.ndarray]
) -> None:
"""
Depending on the model, we might need to preprocess the data before fitting the model.
all required preprocessing and conditional assignments should be defined here.
"""
self.model_coords = None # in our case we're not using coords, but if we were, we would define them here, or later on in the function, if extracting them from the data.
# as we don't do any data preprocessing, we just assign the data given by the user. Note that it's a very basic model,
# and usually we would need to do some preprocessing, or generate the coords from the data.
self.X = X
self.y = y
Now we can create the LinearModel object. First step we need to take care of, is data generation:
X = pd.DataFrame(data=np.linspace(start=0, stop=1, num=100), columns=["input"])
y = 0.3 * x + 0.5
y = y + np.random.normal(0, 1, len(x))
model = LinearModel()
After making the object of class LinearModel we can fit the model using the .fit() method.
Fitting to data#
The fit() method takes one argument data on which we need to fit the model. The meta-data is saved in the InferenceData object where also the trace is stored. These are the fields that are stored:
id: This is a unique id given to a model based on model_config, sample_conifg, version, and model_type. Users can use it to check if the model matches to another model they have defined.model_type: Model type tells us what kind of model it is. This in this case it outputs Linear Modelversion: In case you want to improve on models, you can keep track of model by their version. As the version changes the unique hash in theidalso changes.sample_conifg: It stores values of the sampler configuration set by user for this particular model.model_config: It stores values of the model configuration set by user for this particular model.
idata = model.fit(X, y)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (3 chains in 2 jobs)
NUTS: [a, b, eps]
Sampling 3 chains for 1_000 tune and 1_000 draw iterations (3_000 + 3_000 draws total) took 9 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
Sampling: [a, b, eps, y]
Sampling: [y]
Saving model to file#
After fitting the model, we can probably save it to share the model as a file so one can use it again.
To save() or load(), we can quickly call methods for respective tasks with the following syntax.
fname = "linear_model_v1.nc"
model.save(fname)
This saves a file at the given path, and the name
A NetCDF .nc file that stores the inference data of the model.
Loading a model#
Now if we wanted to deploy this model, or just have other people use it to predict data, they need two things:
the
LinearModelclass (probably in a .py file)the linear_model_v1.nc file
With these, you can easily load a fitted model in a different environment (e.g. production):
model_2 = LinearModel.load(fname)
/media/Data/mambaforge/envs/pymc-docs/lib/python3.11/site-packages/arviz/data/inference_data.py:153: UserWarning: fit_data group is not defined in the InferenceData scheme
warnings.warn(
Note that load() is a class-method, we do not need to instantiate the LinearModel object.
type(model_2)
__main__.LinearModel
Prediction#
Next we might want to predict on new data. The predict() method allows users to do posterior prediction with the fitted model on new data.
Our first task is to create data on which we need to predict.
x_pred = np.random.uniform(low=1, high=2, size=10)
prediction_data = pd.DataFrame({"input": x_pred})
type(prediction_data["input"].values)
numpy.ndarray
ModelBuilder provides two methods for prediction:
point estimates (the mean) with
predict()full posterior prediction (samples) with
predict_posterior()
pred_mean = model_2.predict(prediction_data)
# samples
pred_samples = model_2.predict_posterior(prediction_data)
Sampling: [y]
Sampling: [y]
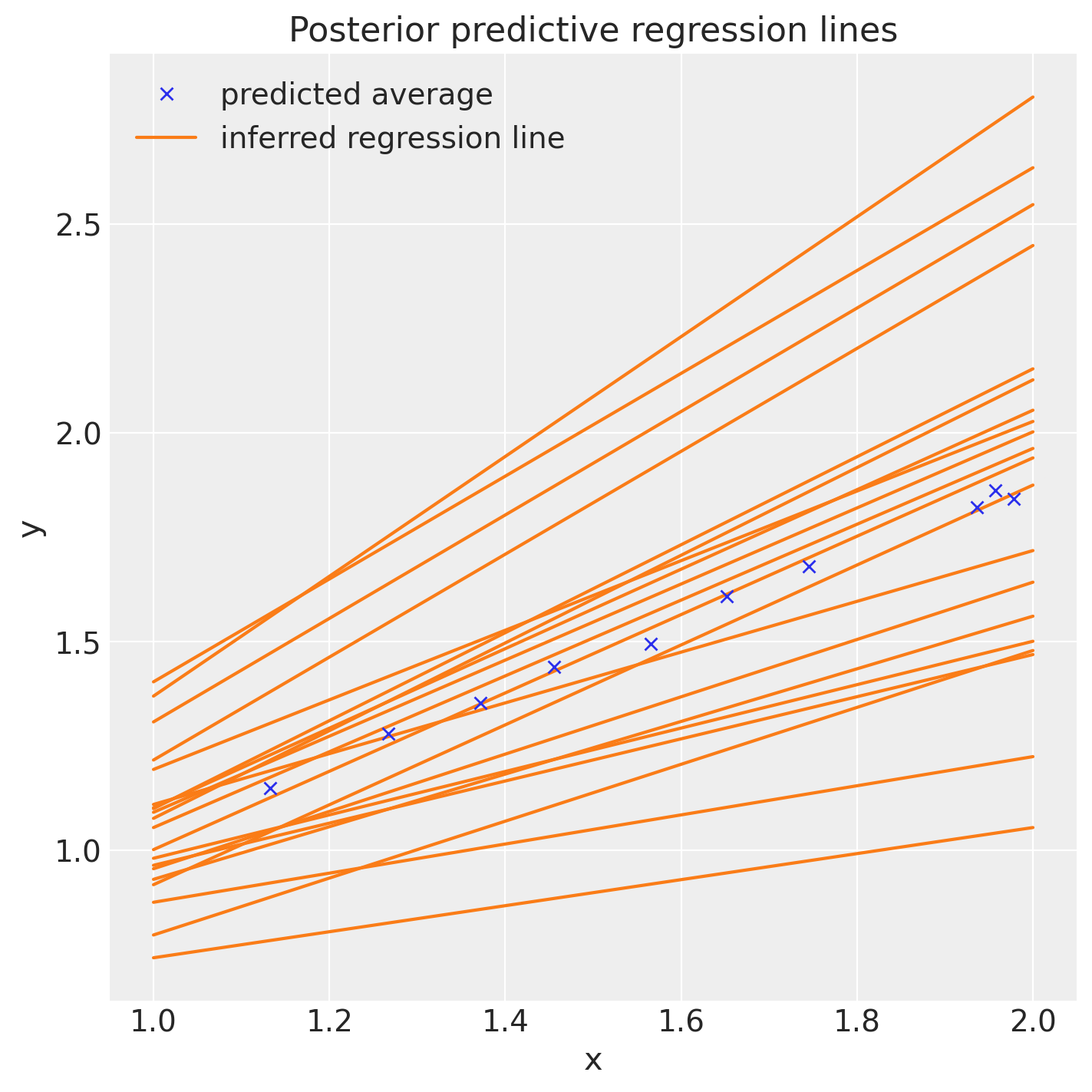
After using the predict(), we can plot our data and see graphically how satisfactory our LinearModel is.
fig, ax = plt.subplots(figsize=(7, 7))
posterior = az.extract(idata, num_samples=20)
x_plot = xr.DataArray(np.linspace(1, 2, 100))
y_plot = posterior["b"] * x_plot + posterior["a"]
Line2 = ax.plot(x_plot, y_plot.transpose(), color="C1")
Line1 = ax.plot(x_pred, pred_mean, "x")
ax.set(title="Posterior predictive regression lines", xlabel="x", ylabel="y")
ax.legend(
handles=[Line1[0], Line2[0]], labels=["predicted average", "inferred regression line"], loc=0
);
%load_ext watermark
%watermark -n -u -v -iv -w -p pymc_experimental
Last updated: Thu Oct 05 2023
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
pymc_experimental: 0.0.12
numpy : 1.25.2
matplotlib: 3.8.0
pandas : 2.1.1
arviz : 0.16.1
xarray : 2023.9.0
pymc : 5.9.0
Watermark: 2.4.3
License notice#
All the notebooks in this example gallery are provided under the MIT License which allows modification, and redistribution for any use provided the copyright and license notices are preserved.
Citing PyMC examples#
To cite this notebook, use the DOI provided by Zenodo for the pymc-examples repository.
Important
Many notebooks are adapted from other sources: blogs, books… In such cases you should cite the original source as well.
Also remember to cite the relevant libraries used by your code.
Here is an citation template in bibtex:
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
which once rendered could look like: