Multi-output Gaussian Processes: Coregionalization models using Hamadard product#
This notebook shows how to implement the Intrinsic Coregionalization Model (ICM) and the Linear Coregionalization Model (LCM) using a Hamadard product between the Coregion kernel and input kernels. Multi-output Gaussian Process is discussed in this paper by Bonilla et al. [2007]. For further information about ICM and LCM, please check out the talk on Multi-output Gaussian Processes by Mauricio Alvarez, and his slides with more references at the last page.
The advantage of Multi-output Gaussian Processes is their capacity to simultaneously learn and infer many outputs which have the same source of uncertainty from inputs. In this example, we model the average spin rates of several pitchers in different games from a baseball dataset.
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc as pm
import pytensor.tensor as pt
from pymc.gp.util import plot_gp_dist
%config InlineBackend.figure_format = 'retina'
RANDOM_SEED = 8927
rng = np.random.default_rng(RANDOM_SEED)
Preparing the data#
The baseball dataset contains the average spin rate of several pitchers on different game dates.
# get data
try:
df = pd.read_csv("../data/fastball_spin_rates.csv")
except FileNotFoundError:
df = pd.read_csv(pm.get_data("fastball_spin_rates.csv"))
print(df.shape)
df.head()
(4845, 4)
| pitcher_name | game_date | avg_spin_rate | n_pitches | |
|---|---|---|---|---|
| 0 | Wainwright, Adam | 2021-04-03 | 2127.415000 | 12 |
| 1 | Wainwright, Adam | 2021-04-08 | 2179.723000 | 11 |
| 2 | Wainwright, Adam | 2021-04-14 | 2297.968571 | 7 |
| 3 | Wainwright, Adam | 2021-04-20 | 2159.150000 | 13 |
| 4 | Wainwright, Adam | 2021-04-26 | 2314.515455 | 11 |
print(
f"There are {df['pitcher_name'].nunique()} pitchers, in {df['game_date'].nunique()} game dates"
)
There are 262 pitchers, in 182 game dates
# Standardise average spin rate
df["avg_spin_rate"] = (df["avg_spin_rate"] - df["avg_spin_rate"].mean()) / df["avg_spin_rate"].std()
df["avg_spin_rate"].describe()
count 4.838000e+03
mean 1.821151e-16
std 1.000000e+00
min -3.646438e+00
25% -6.803556e-01
50% 4.736011e-03
75% 6.837128e-01
max 3.752720e+00
Name: avg_spin_rate, dtype: float64
Top N popular pitchers#
# Get top N popular pitchers by who attended most games
n_outputs = 5 # Top 5 popular pitchers
top_pitchers = df.groupby("pitcher_name")["game_date"].count().nlargest(n_outputs).reset_index()
top_pitchers = top_pitchers.reset_index().rename(columns={"index": "output_idx"})
top_pitchers
| output_idx | pitcher_name | game_date | |
|---|---|---|---|
| 0 | 0 | Rodriguez, Richard | 64 |
| 1 | 1 | Taylor, Josh | 59 |
| 2 | 2 | Kopech, Michael | 43 |
| 3 | 3 | Wells, Tyler | 43 |
| 4 | 4 | Hearn, Taylor | 42 |
# Filter the data with only top N pitchers
adf = df.loc[df["pitcher_name"].isin(top_pitchers["pitcher_name"])].copy()
print(adf.shape)
adf.head()
(251, 4)
| pitcher_name | game_date | avg_spin_rate | n_pitches | |
|---|---|---|---|---|
| 1631 | Rodriguez, Richard | 2021-04-01 | 1.245044 | 12 |
| 1632 | Rodriguez, Richard | 2021-04-06 | 2.032285 | 3 |
| 1633 | Rodriguez, Richard | 2021-04-08 | 1.868068 | 10 |
| 1634 | Rodriguez, Richard | 2021-04-12 | 1.801864 | 14 |
| 1635 | Rodriguez, Richard | 2021-04-13 | 1.916592 | 9 |
adf["avg_spin_rate"].describe()
count 251.000000
mean 0.774065
std 0.867510
min -1.050933
25% 0.102922
50% 0.559293
75% 1.517114
max 2.647740
Name: avg_spin_rate, dtype: float64
Create a game date index#
# There are 142 game dates from 01 Apr 2021 to 03 Oct 2021.
adf.loc[:, "game_date"] = pd.to_datetime(adf.loc[:, "game_date"])
game_dates = adf.loc[:, "game_date"]
game_dates.min(), game_dates.max(), game_dates.nunique(), (game_dates.max() - game_dates.min())
(Timestamp('2021-04-01 00:00:00'),
Timestamp('2021-10-03 00:00:00'),
142,
Timedelta('185 days 00:00:00'))
# Create a game date index
dates_idx = pd.DataFrame(
{"game_date": pd.date_range(game_dates.min(), game_dates.max())}
).reset_index()
dates_idx = dates_idx.rename(columns={"index": "x"})
dates_idx.head()
| x | game_date | |
|---|---|---|
| 0 | 0 | 2021-04-01 |
| 1 | 1 | 2021-04-02 |
| 2 | 2 | 2021-04-03 |
| 3 | 3 | 2021-04-04 |
| 4 | 4 | 2021-04-05 |
Create training data#
adf = adf.merge(dates_idx, how="left", on="game_date")
adf = adf.merge(top_pitchers[["pitcher_name", "output_idx"]], how="left", on="pitcher_name")
adf.head()
| pitcher_name | game_date | avg_spin_rate | n_pitches | x | output_idx | |
|---|---|---|---|---|---|---|
| 0 | Rodriguez, Richard | 2021-04-01 | 1.245044 | 12 | 0 | 0 |
| 1 | Rodriguez, Richard | 2021-04-06 | 2.032285 | 3 | 5 | 0 |
| 2 | Rodriguez, Richard | 2021-04-08 | 1.868068 | 10 | 7 | 0 |
| 3 | Rodriguez, Richard | 2021-04-12 | 1.801864 | 14 | 11 | 0 |
| 4 | Rodriguez, Richard | 2021-04-13 | 1.916592 | 9 | 12 | 0 |
adf = adf.sort_values(["output_idx", "x"])
X = adf[
["x", "output_idx"]
].values # Input data includes the index of game dates, and the index of pitchers
Y = adf["avg_spin_rate"].values # Output data includes the average spin rate of pitchers
X.shape, Y.shape
((251, 2), (251,))
Visualise training data#
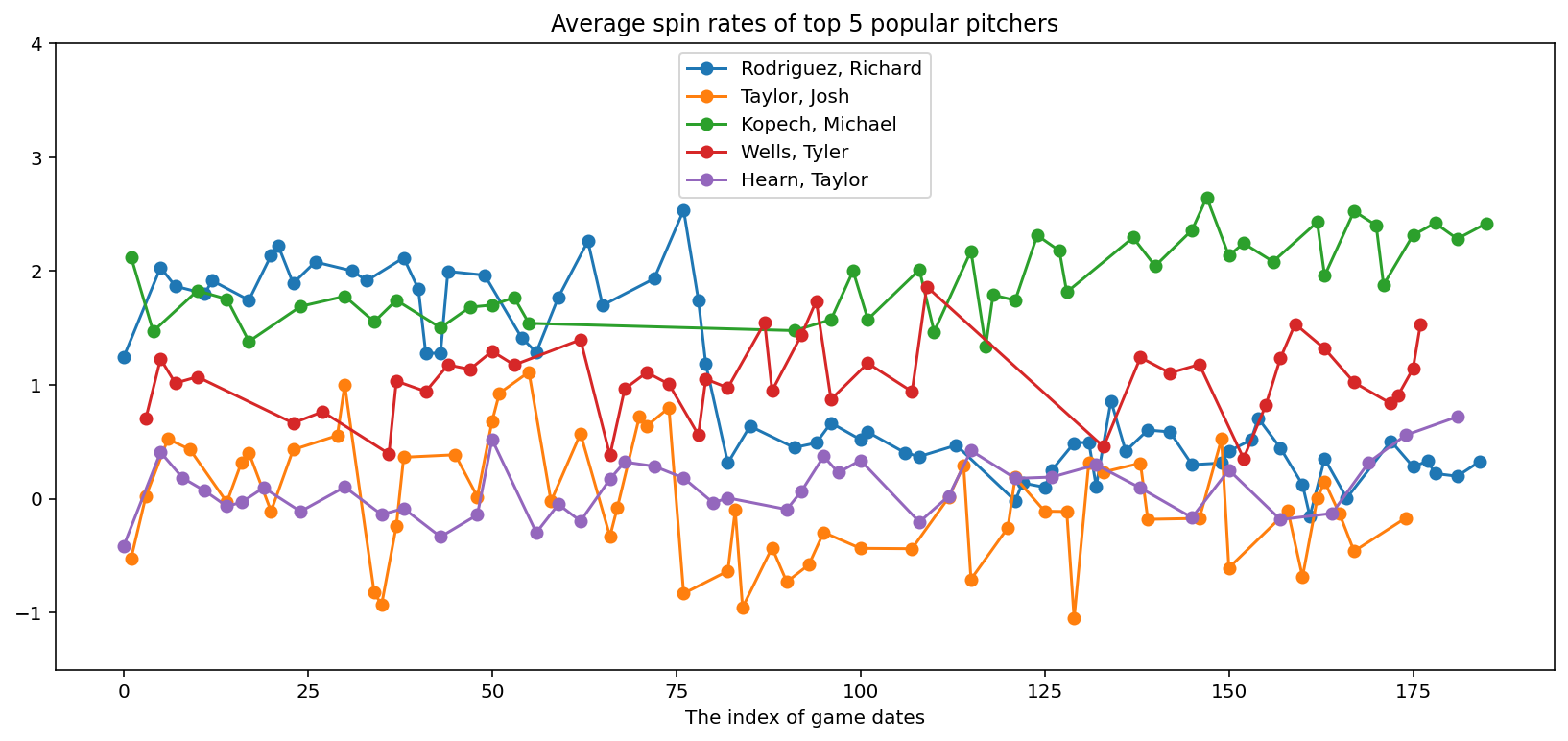
# Plot average spin rates of top pitchers
fig, ax = plt.subplots(1, 1, figsize=(14, 6))
legends = []
for pitcher in top_pitchers["pitcher_name"]:
cond = adf["pitcher_name"] == pitcher
ax.plot(adf.loc[cond, "x"], adf.loc[cond, "avg_spin_rate"], "-o")
legends.append(pitcher)
plt.title("Average spin rates of top 5 popular pitchers")
plt.xlabel("The index of game dates")
plt.ylim([-1.5, 4.0])
plt.legend(legends, loc="upper center");
Intrinsic Coregionalization Model (ICM)#
The Intrinsic Coregionalization Model (ICM) is a particular case of the Linear Coregionalization Model (LCM) with one input kernel, for example:
Where \(B(o,o')\) is the output kernel, and \(K_{ExpQuad}(x,x')\) is an input kernel.
def get_icm(input_dim, kernel, W=None, kappa=None, B=None, active_dims=None):
"""
This function generates an ICM kernel from an input kernel and a Coregion kernel.
"""
coreg = pm.gp.cov.Coregion(input_dim=input_dim, W=W, kappa=kappa, B=B, active_dims=active_dims)
icm_cov = kernel * coreg # Use Hadamard Product for separate inputs
return icm_cov
with pm.Model() as model:
# Priors
ell = pm.Gamma("ell", alpha=2, beta=0.5)
eta = pm.Gamma("eta", alpha=3, beta=1)
kernel = eta**2 * pm.gp.cov.ExpQuad(input_dim=2, ls=ell, active_dims=[0])
sigma = pm.HalfNormal("sigma", sigma=3)
# Get the ICM kernel
W = pm.Normal("W", mu=0, sigma=3, shape=(n_outputs, 2), initval=np.random.randn(n_outputs, 2))
kappa = pm.Gamma("kappa", alpha=1.5, beta=1, shape=n_outputs)
B = pm.Deterministic("B", pt.dot(W, W.T) + pt.diag(kappa))
cov_icm = get_icm(input_dim=2, kernel=kernel, B=B, active_dims=[1])
# Define a Multi-output GP
mogp = pm.gp.Marginal(cov_func=cov_icm)
y_ = mogp.marginal_likelihood("f", X, Y, sigma=sigma)
pm.model_to_graphviz(model)
%%time
with model:
gp_trace = pm.sample(2000, chains=1)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (1 chains in 1 job)
NUTS: [ell, eta, sigma, W, kappa]
Sampling 1 chain for 1_000 tune and 2_000 draw iterations (1_000 + 2_000 draws total) took 1278 seconds.
CPU times: user 46min 50s, sys: 2h 2min 57s, total: 2h 49min 47s
Wall time: 21min 25s
Prediction#
# Prepare test data
M = 200 # number of data points
x_new = np.linspace(0, 200, M)[
:, None
] # Select 200 days (185 previous days, and add 15 days into the future).
X_new = np.vstack([x_new for idx in range(n_outputs)])
output_idx = np.vstack([np.repeat(idx, M)[:, None] for idx in range(n_outputs)])
X_new = np.hstack([X_new, output_idx])
%%time
with model:
preds = mogp.conditional("preds", X_new)
gp_samples = pm.sample_posterior_predictive(gp_trace, var_names=["preds"], random_seed=42)
Sampling: [preds]
CPU times: user 46min 13s, sys: 39min 48s, total: 1h 26min 2s
Wall time: 10min 50s
f_pred = gp_samples.posterior_predictive["preds"].sel(chain=0)
def plot_predictive_posteriors(f_pred, top_pitchers, M, X_new):
fig, axes = plt.subplots(n_outputs, 1, figsize=(12, 15))
for idx, pitcher in enumerate(top_pitchers["pitcher_name"]):
# Prediction
plot_gp_dist(
axes[idx],
f_pred[:, M * idx : M * (idx + 1)],
X_new[M * idx : M * (idx + 1), 0],
palette="Blues",
fill_alpha=0.1,
samples_alpha=0.1,
)
# Training data points
cond = adf["pitcher_name"] == pitcher
axes[idx].scatter(adf.loc[cond, "x"], adf.loc[cond, "avg_spin_rate"], color="r")
axes[idx].set_title(pitcher)
plt.tight_layout()
plot_predictive_posteriors(f_pred, top_pitchers, M, X_new)
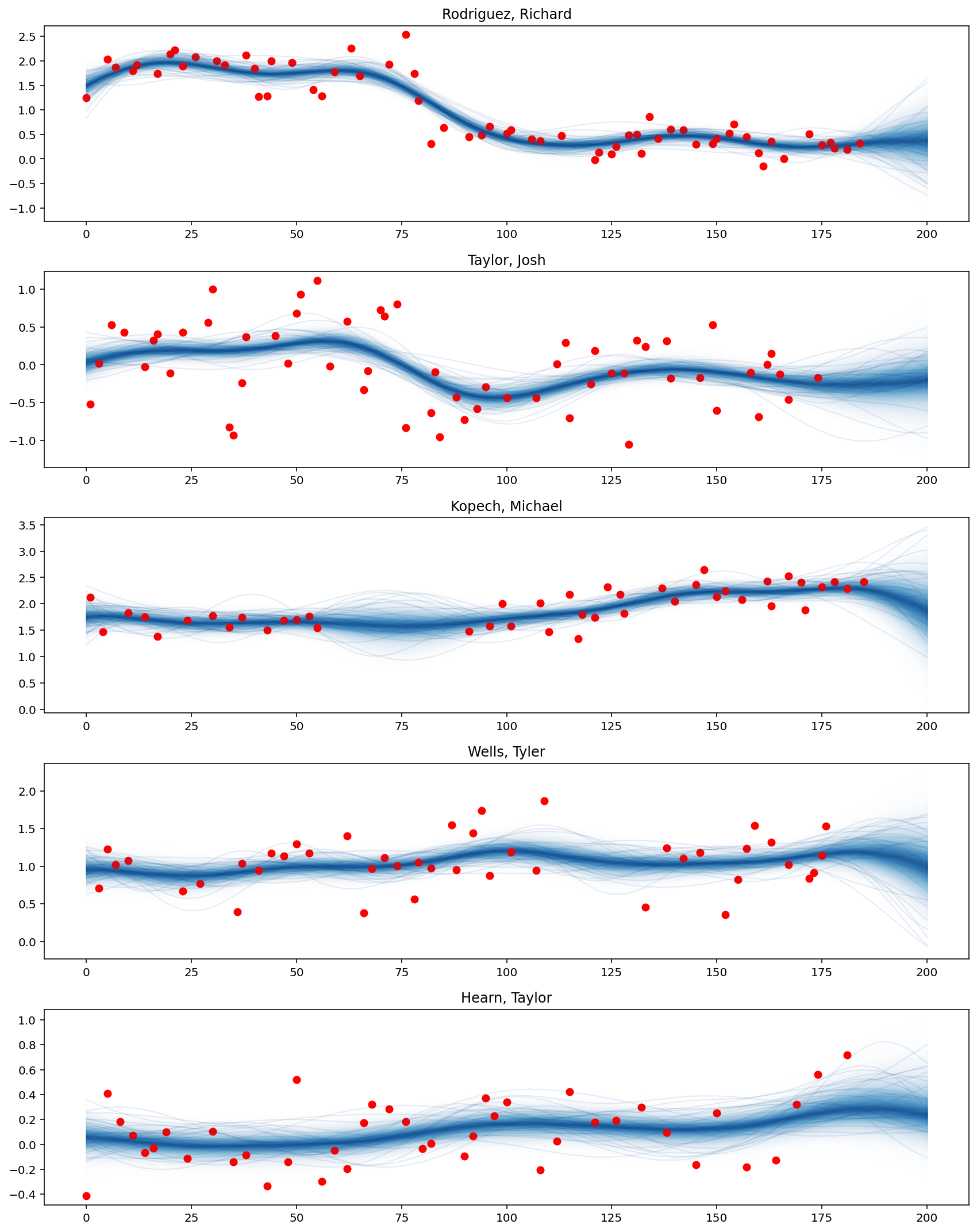
It can be seen that the average spin rate of Rodriguez Richard decreases significantly from the 75th game dates. Besides, Kopech Michael’s performance improves after a break of several weeks in the middle, while Hearn Taylor has performed better recently.
az.plot_trace(gp_trace)
plt.tight_layout()
Linear Coregionalization Model (LCM)#
The LCM is a generalization of the ICM with two or more input kernels, so the LCM kernel is basically a sum of several ICM kernels. The LMC allows several independent samples from GPs with different covariances (kernels).
In this example, in addition to an ExpQuad kernel, we add a Matern32 kernel for input data.
def get_lcm(input_dim, active_dims, num_outputs, kernels, W=None, B=None, name="ICM"):
"""
This function generates a LCM kernel from a list of input `kernels` and a Coregion kernel.
"""
if B is None:
kappa = pm.Gamma(f"{name}_kappa", alpha=5, beta=1, shape=num_outputs)
if W is None:
W = pm.Normal(
f"{name}_W",
mu=0,
sigma=5,
shape=(num_outputs, 1),
initval=np.random.randn(num_outputs, 1),
)
else:
kappa = None
cov_func = 0
for idx, kernel in enumerate(kernels):
icm = get_icm(input_dim, kernel, W, kappa, B, active_dims)
cov_func += icm
return cov_func
with pm.Model() as model:
# Priors
ell = pm.Gamma("ell", alpha=2, beta=0.5, shape=2)
eta = pm.Gamma("eta", alpha=3, beta=1, shape=2)
kernels = [pm.gp.cov.ExpQuad, pm.gp.cov.Matern32]
sigma = pm.HalfNormal("sigma", sigma=3)
# Define a list of covariance functions
cov_list = [
eta[idx] ** 2 * kernel(input_dim=2, ls=ell[idx], active_dims=[0])
for idx, kernel in enumerate(kernels)
]
# Get the LCM kernel
cov_lcm = get_lcm(input_dim=2, active_dims=[1], num_outputs=n_outputs, kernels=cov_list)
# Define a Multi-output GP
mogp = pm.gp.Marginal(cov_func=cov_lcm)
y_ = mogp.marginal_likelihood("f", X, Y, sigma=sigma)
pm.model_to_graphviz(model)
%%time
with model:
gp_trace = pm.sample(2000, chains=1)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (1 chains in 1 job)
NUTS: [ell, eta, sigma, ICM_kappa, ICM_W]
Sampling 1 chain for 1_000 tune and 2_000 draw iterations (1_000 + 2_000 draws total) took 1109 seconds.
There were 2 divergences after tuning. Increase `target_accept` or reparameterize.
CPU times: user 40min 37s, sys: 1h 47min 9s, total: 2h 27min 46s
Wall time: 18min 39s
Prediction#
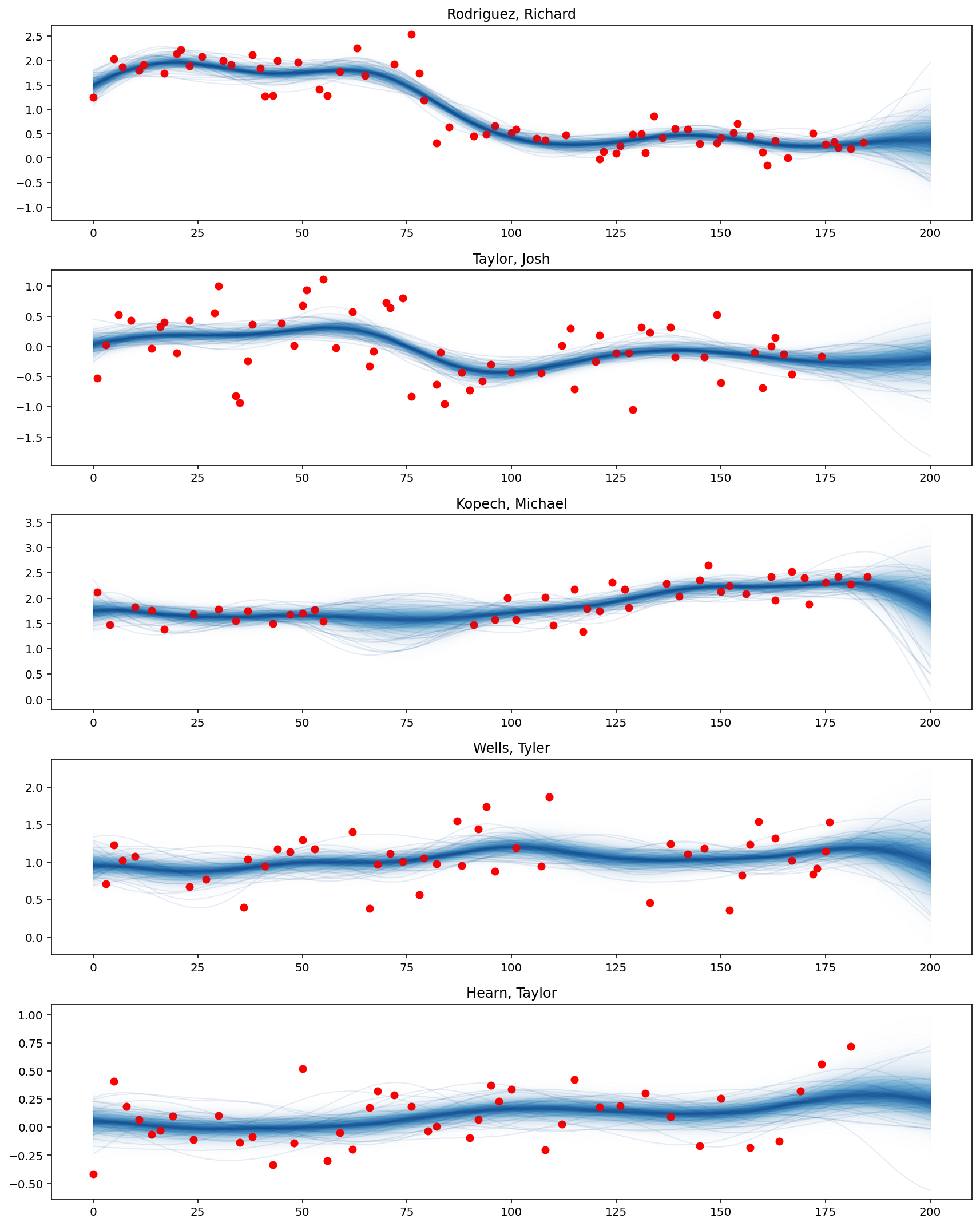
%%time
with model:
preds = mogp.conditional("preds", X_new)
gp_samples = pm.sample_posterior_predictive(gp_trace, var_names=["preds"], random_seed=42)
Sampling: [preds]
CPU times: user 51min 44s, sys: 50min 14s, total: 1h 41min 59s
Wall time: 12min 53s
plot_predictive_posteriors(f_pred, top_pitchers, M, X_new)
az.plot_trace(gp_trace)
plt.tight_layout()
Acknowledgement#
This work is supported by 2022 Google Summer of Codes and NUMFOCUS.
References#
Edwin V Bonilla, Kian Chai, and Christopher Williams. Multi-task gaussian process prediction. Advances in neural information processing systems, 2007. URL: https://papers.nips.cc/paper/2007/hash/66368270ffd51418ec58bd793f2d9b1b-Abstract.html.
Watermark#
%load_ext watermark
%watermark -n -u -v -iv -w -p pytensor,aeppl,xarray
The watermark extension is already loaded. To reload it, use:
%reload_ext watermark
Last updated: Sat Nov 12 2022
Python implementation: CPython
Python version : 3.9.12
IPython version : 8.3.0
pytensor: 2.8.6
aeppl : 0.0.36
xarray: 2022.3.0
pymc : 4.2.1
arviz : 0.13.0
pandas : 1.4.2
pytensor : 2.8.6
numpy : 1.22.4
matplotlib: 3.5.2
Watermark: 2.3.0
License notice#
All the notebooks in this example gallery are provided under the MIT License which allows modification, and redistribution for any use provided the copyright and license notices are preserved.
Citing PyMC examples#
To cite this notebook, use the DOI provided by Zenodo for the pymc-examples repository.
Important
Many notebooks are adapted from other sources: blogs, books… In such cases you should cite the original source as well.
Also remember to cite the relevant libraries used by your code.
Here is an citation template in bibtex:
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
which once rendered could look like: