GLM: Mini-batch ADVI on hierarchical regression model#
Unlike Gaussian mixture models, (hierarchical) regression models have independent variables. These variables affect the likelihood function, but are not random variables. When using mini-batch, we should take care of that.
%env PYTENSOR_FLAGS=device=cpu, floatX=float32, warn_float64=ignore
import os
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc as pm
import pytensor
import pytensor.tensor as pt
import seaborn as sns
from scipy import stats
print(f"Running on PyMC v{pm.__version__}")
env: PYTENSOR_FLAGS=device=cpu, floatX=float32, warn_float64=ignore
Running on PyMC v5.0.1
%config InlineBackend.figure_format = 'retina'
RANDOM_SEED = 8927
rng = np.random.default_rng(RANDOM_SEED)
az.style.use("arviz-darkgrid")
try:
data = pd.read_csv(os.path.join("..", "data", "radon.csv"))
except FileNotFoundError:
data = pd.read_csv(pm.get_data("radon.csv"))
data
| Unnamed: 0 | idnum | state | state2 | stfips | zip | region | typebldg | floor | room | ... | pcterr | adjwt | dupflag | zipflag | cntyfips | county | fips | Uppm | county_code | log_radon | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 5081.0 | MN | MN | 27.0 | 55735 | 5.0 | 1.0 | 1.0 | 3.0 | ... | 9.7 | 1146.499190 | 1.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 0.832909 |
| 1 | 1 | 5082.0 | MN | MN | 27.0 | 55748 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 14.5 | 471.366223 | 0.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 0.832909 |
| 2 | 2 | 5083.0 | MN | MN | 27.0 | 55748 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 9.6 | 433.316718 | 0.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 1.098612 |
| 3 | 3 | 5084.0 | MN | MN | 27.0 | 56469 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 24.3 | 461.623670 | 0.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 0.095310 |
| 4 | 4 | 5085.0 | MN | MN | 27.0 | 55011 | 3.0 | 1.0 | 0.0 | 4.0 | ... | 13.8 | 433.316718 | 0.0 | 0.0 | 3.0 | ANOKA | 27003.0 | 0.428565 | 1 | 1.163151 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 914 | 914 | 5995.0 | MN | MN | 27.0 | 55363 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 4.5 | 1146.499190 | 0.0 | 0.0 | 171.0 | WRIGHT | 27171.0 | 0.913909 | 83 | 1.871802 |
| 915 | 915 | 5996.0 | MN | MN | 27.0 | 55376 | 5.0 | 1.0 | 0.0 | 7.0 | ... | 8.3 | 1105.956867 | 0.0 | 0.0 | 171.0 | WRIGHT | 27171.0 | 0.913909 | 83 | 1.526056 |
| 916 | 916 | 5997.0 | MN | MN | 27.0 | 55376 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 5.2 | 1214.922779 | 0.0 | 0.0 | 171.0 | WRIGHT | 27171.0 | 0.913909 | 83 | 1.629241 |
| 917 | 917 | 5998.0 | MN | MN | 27.0 | 56297 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 9.6 | 1177.377355 | 0.0 | 0.0 | 173.0 | YELLOW MEDICINE | 27173.0 | 1.426590 | 84 | 1.335001 |
| 918 | 918 | 5999.0 | MN | MN | 27.0 | 56297 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 8.0 | 1214.922779 | 0.0 | 0.0 | 173.0 | YELLOW MEDICINE | 27173.0 | 1.426590 | 84 | 1.098612 |
919 rows × 30 columns
county_idx = data["county_code"].values
floor_idx = data["floor"].values
log_radon_idx = data["log_radon"].values
coords = {"counties": data.county.unique()}
Here, log_radon_idx_t is a dependent variable, while floor_idx_t and county_idx_t determine the independent variable.
log_radon_idx_t = pm.Minibatch(log_radon_idx, batch_size=100)
floor_idx_t = pm.Minibatch(floor_idx, batch_size=100)
county_idx_t = pm.Minibatch(county_idx, batch_size=100)
with pm.Model(coords=coords) as hierarchical_model:
# Hyperpriors for group nodes
mu_a = pm.Normal("mu_alpha", mu=0.0, sigma=100**2)
sigma_a = pm.Uniform("sigma_alpha", lower=0, upper=100)
mu_b = pm.Normal("mu_beta", mu=0.0, sigma=100**2)
sigma_b = pm.Uniform("sigma_beta", lower=0, upper=100)
Intercept for each county, distributed around group mean mu_a. Above we just set mu and sd to a fixed value while here we plug in a common group distribution for all a and b (which are vectors with the same length as the number of unique counties in our example).
Model prediction of radon level a[county_idx] translates to a[0, 0, 0, 1, 1, ...], we thus link multiple household measures of a county to its coefficients.
with hierarchical_model:
radon_est = a[county_idx_t] + b[county_idx_t] * floor_idx_t
Finally, we specify the likelihood:
with hierarchical_model:
# Model error
eps = pm.Uniform("eps", lower=0, upper=100)
# Data likelihood
radon_like = pm.Normal(
"radon_like", mu=radon_est, sigma=eps, observed=log_radon_idx_t, total_size=len(data)
)
Random variables radon_like, associated with log_radon_idx_t, should be given to the function for ADVI to denote that as observations in the likelihood term.
On the other hand, minibatches should include the three variables above.
Then, run ADVI with mini-batch.
with hierarchical_model:
approx = pm.fit(100_000, callbacks=[pm.callbacks.CheckParametersConvergence(tolerance=1e-4)])
idata_advi = approx.sample(500)
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
Check the trace of ELBO and compare the result with MCMC.
plt.plot(approx.hist);
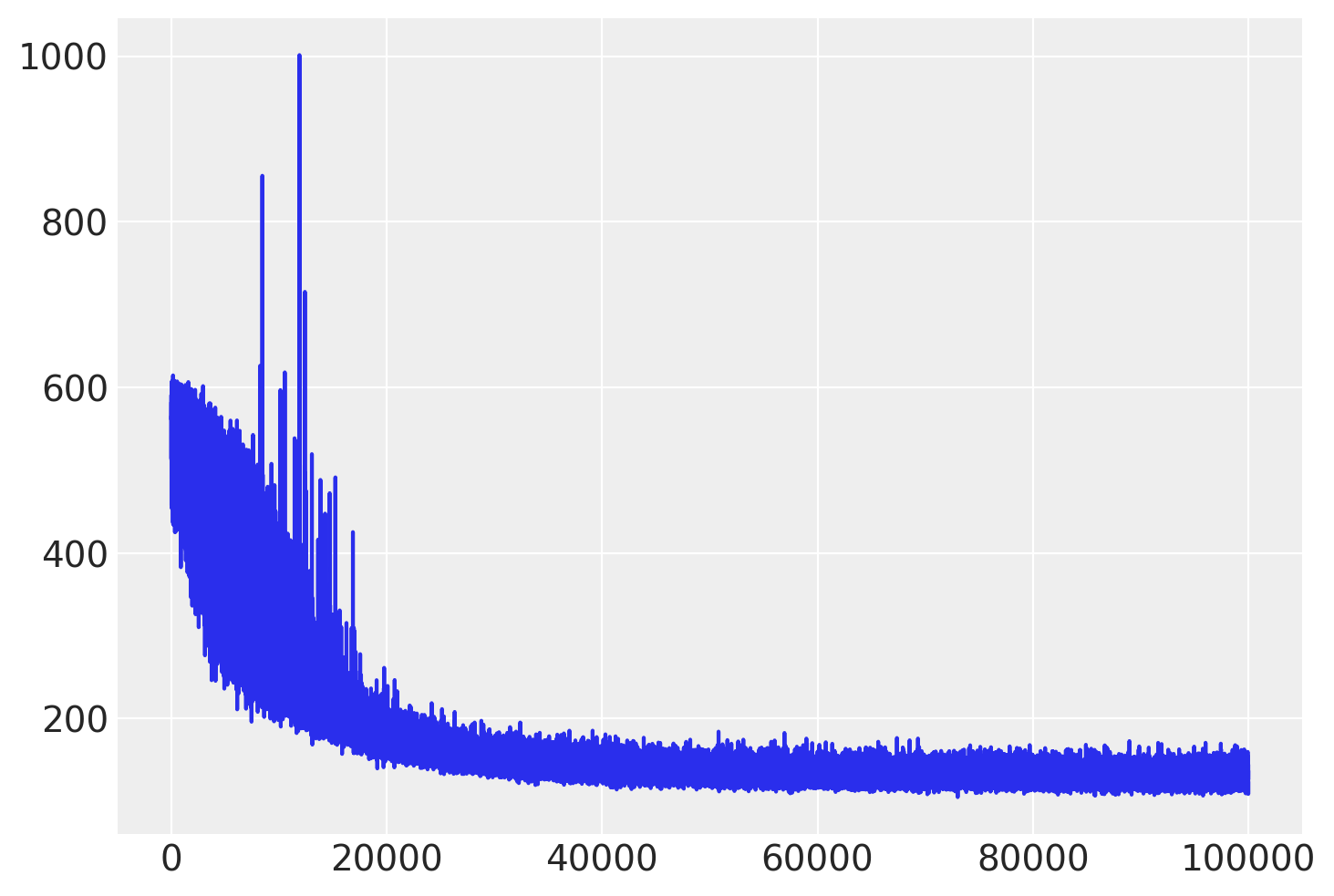
We can extract the covariance matrix from the mean field approximation and use it as the scaling matrix for the NUTS algorithm.
scaling = approx.cov.eval()
Also, we can generate samples (one for each chain) to use as the starting points for the sampler.
n_chains = 4
sample = approx.sample(return_inferencedata=False, size=n_chains)
start_dict = list(sample[i] for i in range(n_chains))
# Inference button (TM)!
with pm.Model(coords=coords):
mu_a = pm.Normal("mu_alpha", mu=0.0, sigma=100**2)
sigma_a = pm.Uniform("sigma_alpha", lower=0, upper=100)
mu_b = pm.Normal("mu_beta", mu=0.0, sigma=100**2)
sigma_b = pm.Uniform("sigma_beta", lower=0, upper=100)
a = pm.Normal("alpha", mu=mu_a, sigma=sigma_a, dims="counties")
b = pm.Normal("beta", mu=mu_b, sigma=sigma_b, dims="counties")
# Model error
eps = pm.Uniform("eps", lower=0, upper=100)
radon_est = a[county_idx] + b[county_idx] * floor_idx
radon_like = pm.Normal("radon_like", mu=radon_est, sigma=eps, observed=log_radon_idx)
# essentially, this is what init='advi' does
step = pm.NUTS(scaling=scaling, is_cov=True)
hierarchical_trace = pm.sample(draws=2000, step=step, chains=n_chains, initvals=start_dict)
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [mu_alpha, sigma_alpha, mu_beta, sigma_beta, alpha, beta, eps]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 538 seconds.
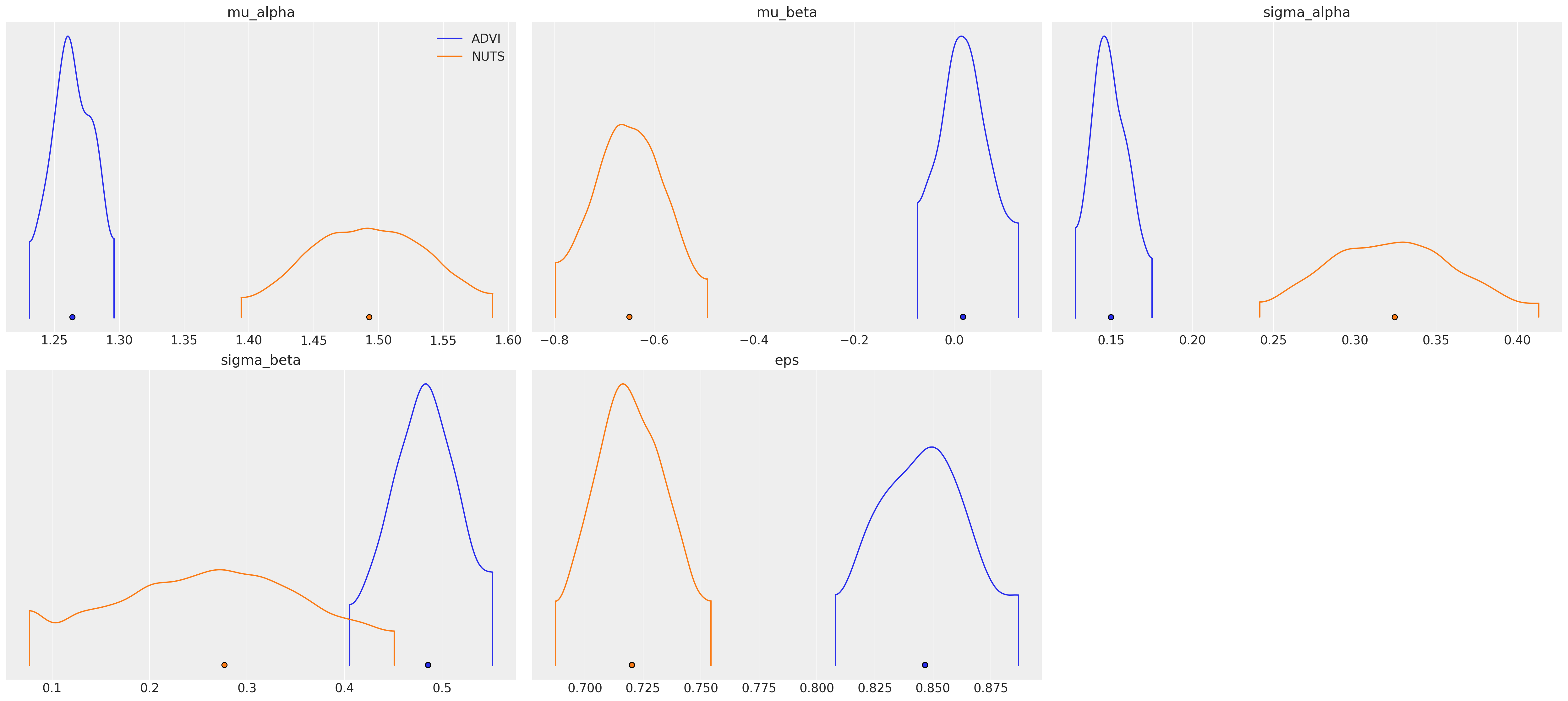
az.plot_density(
data=[idata_advi, hierarchical_trace],
var_names=["~alpha", "~beta"],
data_labels=["ADVI", "NUTS"],
);
Watermark#
%load_ext watermark
%watermark -n -u -v -iv -w -p xarray
Last updated: Thu Sep 23 2021
Python implementation: CPython
Python version : 3.8.10
IPython version : 7.25.0
xarray: 0.17.0
numpy : 1.21.0
seaborn : 0.11.1
pandas : 1.2.1
matplotlib: 3.3.4
theano : 1.1.2
pymc3 : 3.11.2
scipy : 1.7.1
arviz : 0.11.2
Watermark: 2.2.0
License notice#
All the notebooks in this example gallery are provided under the MIT License which allows modification, and redistribution for any use provided the copyright and license notices are preserved.
Citing PyMC examples#
To cite this notebook, use the DOI provided by Zenodo for the pymc-examples repository.
Important
Many notebooks are adapted from other sources: blogs, books… In such cases you should cite the original source as well.
Also remember to cite the relevant libraries used by your code.
Here is an citation template in bibtex:
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
which once rendered could look like: