Multivariate Gaussian Random Walk¶
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pymc3 as pm
import theano
from scipy.linalg import cholesky
%matplotlib inline
RANDOM_SEED = 8927
rng = np.random.default_rng(RANDOM_SEED)
az.style.use("arviz-darkgrid")
This notebook shows how to fit a correlated time series using multivariate Gaussian random walks (GRWs). In particular, we perform a Bayesian regression of the time series data against a model dependent on GRWs.
We generate data as the 3-dimensional time series
where
\(i\mapsto\alpha_{i}\) and \(i\mapsto\beta_{i}\), \(i\in\{0,1,2,3,4\}\), are two 3-dimensional Gaussian random walks for two correlation matrices \(\Sigma_\alpha\) and \(\Sigma_\beta\),
we define the index $\( i[t]= j\quad\text{for}\quad t = 60j,60j+1,...,60j+59, \quad\text{and}\quad j = 0,1,2,3,4, \)$
\(*\) means that we multiply the \(j\)-th column of the \(3\times300\) matrix with the \(j\)-th entry of the vector for each \(j=0,1,...,299\), and
\(\xi_{\mathbf t}\) is a \(3\times300\) matrix with iid normal entries \(N(0,\sigma^2)\).
So the series \(\mathbf y\) changes due to the GRW \(\alpha\) in five occasions, namely steps \(0,60,120,180,240\). Meanwhile \(\mathbf y\) changes at steps \(1,60,120,180,240\) due to the increments of the GRW \(\beta\) and at every step due to the weighting of \(\beta\) with \(\mathbf t/300\). Intuitively, we have a noisy (\(\xi\)) system that is shocked five times over a period of 300 steps, but the impact of the \(\beta\) shocks gradually becomes more significant at every step.
Data generation¶
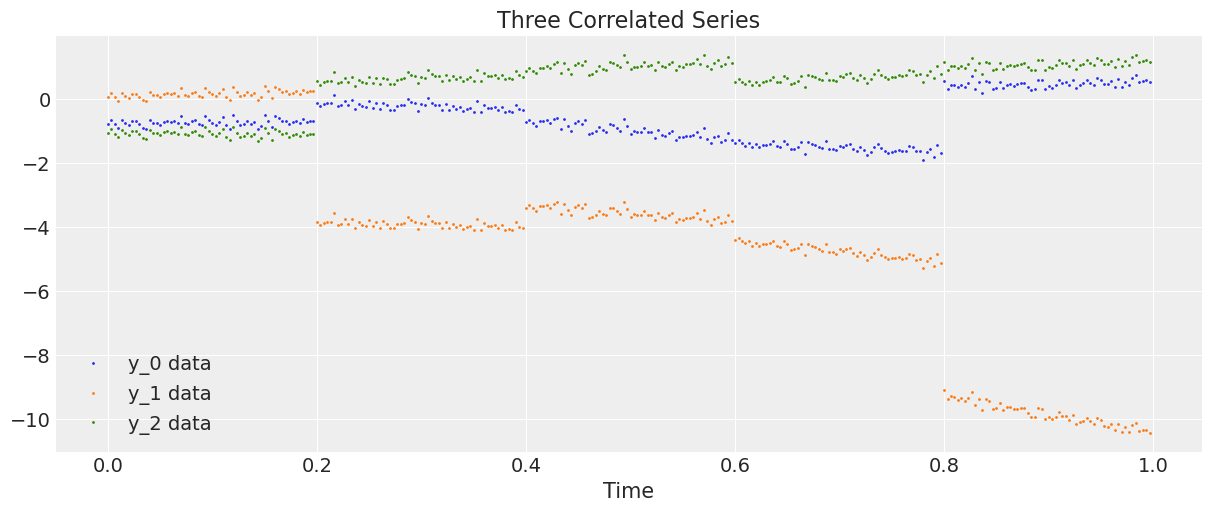
Let’s generate and plot the data.
D = 3 # Dimension of random walks
N = 300 # Number of steps
sections = 5 # Number of sections
period = N / sections # Number steps in each section
Sigma_alpha = rng.standard_normal((D, D))
Sigma_alpha = Sigma_alpha.T.dot(Sigma_alpha) # Construct covariance matrix for alpha
L_alpha = cholesky(Sigma_alpha, lower=True) # Obtain its Cholesky decomposition
Sigma_beta = rng.standard_normal((D, D))
Sigma_beta = Sigma_beta.T.dot(Sigma_beta) # Construct covariance matrix for beta
L_beta = cholesky(Sigma_beta, lower=True) # Obtain its Cholesky decomposition
# Gaussian random walks:
alpha = np.cumsum(L_alpha.dot(rng.standard_normal((D, sections))), axis=1).T
beta = np.cumsum(L_beta.dot(rng.standard_normal((D, sections))), axis=1).T
t = np.arange(N)[:, None] / N
alpha = np.repeat(alpha, period, axis=0)
beta = np.repeat(beta, period, axis=0)
# Correlated series
sigma = 0.1
y = alpha + beta * t + sigma * rng.standard_normal((N, 1))
# Plot the correlated series
plt.figure(figsize=(12, 5))
plt.plot(t, y, ".", markersize=2, label=("y_0 data", "y_1 data", "y_2 data"))
plt.title("Three Correlated Series")
plt.xlabel("Time")
plt.legend()
plt.show();
Model¶
First we introduce a scaling class to rescale our data and the time parameter before the sampling and then rescale the predictions to match the unscaled data.
class Scaler:
def __init__(self):
mean_ = None
std_ = None
def transform(self, x):
return (x - self.mean_) / self.std_
def fit_transform(self, x):
self.mean_ = x.mean(axis=0)
self.std_ = x.std(axis=0)
return self.transform(x)
def inverse_transform(self, x):
return x * self.std_ + self.mean_
We now construct the regression model in (1) imposing priors on the GRWs \(\alpha\) and \(\beta\), on the standard deviation \(\sigma\) and hyperpriors on the Cholesky matrices. We use the LKJ prior [Lewandowski et al., 2009] for the Cholesky matrices (see this link for the documentation and also the PyMC notebook LKJ Cholesky Covariance Priors for Multivariate Normal Models for some usage examples.)
def inference(t, y, sections, n_samples=100):
N, D = y.shape
# Standardies y and t
y_scaler = Scaler()
t_scaler = Scaler()
y = y_scaler.fit_transform(y)
t = t_scaler.fit_transform(t)
# Create a section index
t_section = np.repeat(np.arange(sections), N / sections)
# Create theano equivalent
t_t = theano.shared(np.repeat(t, D, axis=1))
y_t = theano.shared(y)
t_section_t = theano.shared(t_section)
coords = {"y_": ["y_0", "y_1", "y_2"], "steps": np.arange(N)}
with pm.Model(coords=coords) as model:
# Hyperpriors on Cholesky matrices
packed_L_alpha = pm.LKJCholeskyCov(
"packed_L_alpha", n=D, eta=2.0, sd_dist=pm.HalfCauchy.dist(2.5)
)
L_alpha = pm.expand_packed_triangular(D, packed_L_alpha)
packed_L_beta = pm.LKJCholeskyCov(
"packed_L_beta", n=D, eta=2.0, sd_dist=pm.HalfCauchy.dist(2.5)
)
L_beta = pm.expand_packed_triangular(D, packed_L_beta)
# Priors on Gaussian random walks
alpha = pm.MvGaussianRandomWalk("alpha", shape=(sections, D), chol=L_alpha)
beta = pm.MvGaussianRandomWalk("beta", shape=(sections, D), chol=L_beta)
# Deterministic construction of the correlated random walk
alpha_r = alpha[t_section_t]
beta_r = beta[t_section_t]
regression = alpha_r + beta_r * t_t
# Prior on noise ξ
sigma = pm.HalfNormal("sigma", 1.0)
# Likelihood
likelihood = pm.Normal("y", mu=regression, sigma=sigma, observed=y_t, dims=("steps", "y_"))
# MCMC sampling
trace = pm.sample(n_samples, cores=4, return_inferencedata=True)
# Posterior predictive sampling
trace.extend(az.from_pymc3(posterior_predictive=pm.sample_posterior_predictive(trace)))
return trace, y_scaler, t_scaler, t_section
Inference¶
We now sample from our model and we return the trace, the scaling functions for space and time and the scaled time index.
trace, y_scaler, t_scaler, t_section = inference(t, y, sections)
Only 100 samples in chain.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [sigma, beta, alpha, packed_L_beta, packed_L_alpha]
Sampling 4 chains for 1_000 tune and 100 draw iterations (4_000 + 400 draws total) took 1836 seconds.
0, dim: steps, 300 =? 300
1, dim: y_, 3 =? 3
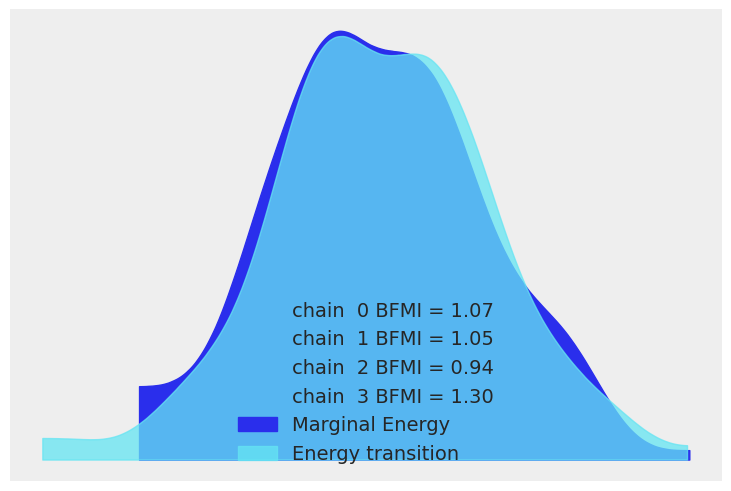
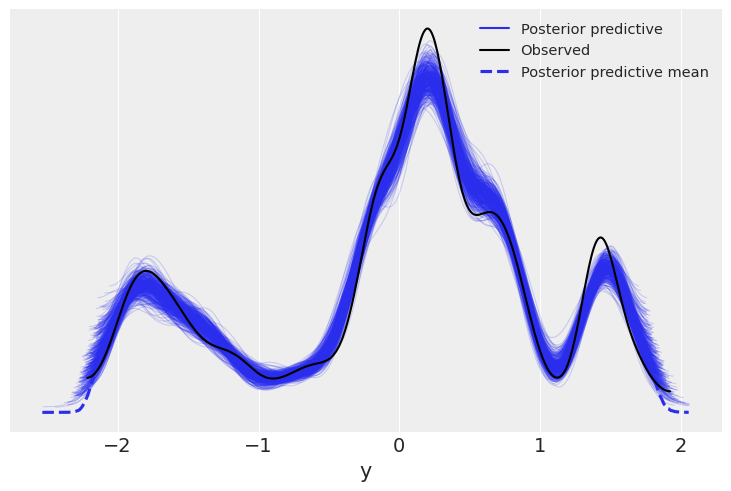
We now display the energy plot using arviz.plot_energy() for a visual check for the model’s convergence. Then, using arviz.plot_ppc(), we plot the distribution of the posterior predictive samples against the observed data \(\mathbf y\). This plot provides a general idea of the accuracy of the model (note that the values of \(\mathbf y\) actually correspond to the scaled version of \(\mathbf y\)).
az.plot_energy(trace)
az.plot_ppc(trace);
Posterior visualisation¶
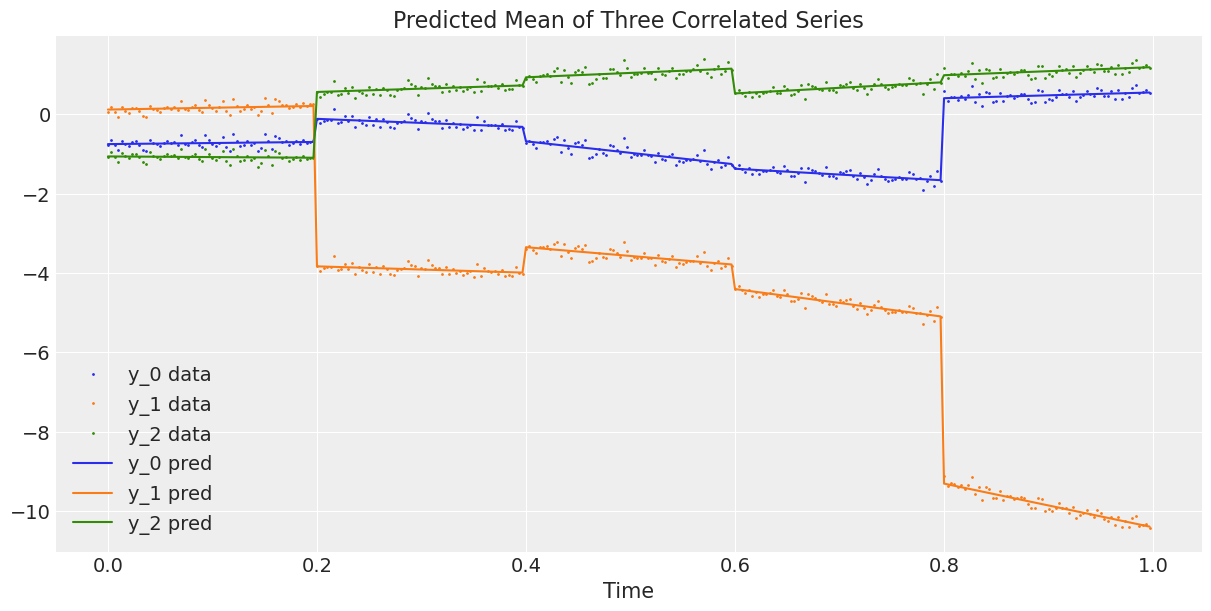
The graphs above look good. Now we plot the observed 3-dimensional series against the average predicted 3-dimensional series, or in other words, we plot the data against the estimated regression curve from the model (1).
# Compute the predicted mean of the multivariate GRWs
alpha_mean = trace.posterior["alpha"].mean(dim=("chain", "draw"))
beta_mean = trace.posterior["beta"].mean(dim=("chain", "draw"))
# Compute the predicted mean of the correlated series
y_pred = y_scaler.inverse_transform(
alpha_mean[t_section].values + beta_mean[t_section].values * t_scaler.transform(t)
)
# Plot the predicted mean
fig, ax = plt.subplots(1, 1, figsize=(12, 6))
ax.plot(t, y, ".", markersize=2, label=("y_0 data", "y_1 data", "y_2 data"))
plt.gca().set_prop_cycle(None)
ax.plot(t, y_pred, label=("y_0 pred", "y_1 pred", "y_2 pred"))
ax.set_xlabel("Time")
ax.legend()
ax.set_title("Predicted Mean of Three Correlated Series");
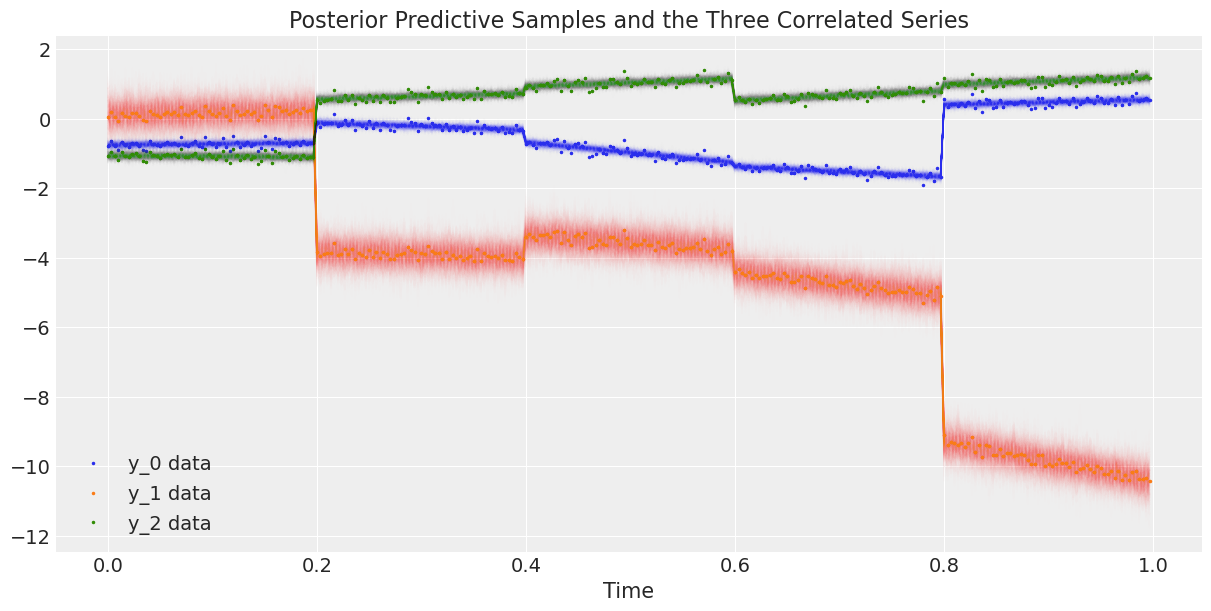
Finally, we plot the data against the posterior predictive samples.
# Rescale the posterior predictive samples
ppc_y = y_scaler.inverse_transform(trace.posterior_predictive["y"].mean("chain"))
fig, ax = plt.subplots(1, 1, figsize=(12, 6))
# Plot the data
ax.plot(t, y, ".", markersize=3, label=("y_0 data", "y_1 data", "y_2 data"))
# Plot the posterior predictive samples
ax.plot(t, ppc_y.sel(y_="y_0").T, color="C0", alpha=0.003)
ax.plot(t, ppc_y.sel(y_="y_1").T, color="C1", alpha=0.003)
ax.plot(t, ppc_y.sel(y_="y_2").T, color="C2", alpha=0.003)
ax.set_xlabel("Time")
ax.legend()
ax.set_title("Posterior Predictive Samples and the Three Correlated Series");
References¶
- 1
Daniel Lewandowski, Dorota Kurowicka, and Harry Joe. Generating random correlation matrices based on vines and extended onion method. Journal of multivariate analysis, 100(9):1989–2001, 2009.
Watermark¶
%load_ext watermark
%watermark -n -u -v -iv -w -p theano,xarray
Last updated: Sat Sep 25 2021
Python implementation: CPython
Python version : 3.9.6
IPython version : 7.26.0
theano: 1.0.5
xarray: 0.19.0
arviz : 0.11.2
numpy : 1.21.1
matplotlib: 3.4.2
theano : 1.0.5
pymc3 : 3.11.2
Watermark: 2.2.0
- PyMC Contributors . "Multivariate Gaussian Random Walk". In: PyMC Examples. Ed. by PyMC Team. DOI: 10.5281/zenodo.5832070