Introductory Overview of PyMC#
Note: This text is partly based on the PeerJ CS publication on PyMC by John Salvatier, Thomas V. Wiecki, and Christopher Fonnesbeck.
Abstract#
Probabilistic Programming allows for automatic Bayesian inference on user-defined probabilistic models. Gradient-based algorithms for Markov chain Monte Carlo (MCMC) sampling, known as Hamiltonian Monte Carlo (HMC), allow inference on increasingly complex models but requires gradient information that is often not trivial to calculate. PyMC is an open source probabilistic programming framework written in Python that uses PyTensor to compute gradients via automatic differentiation, as well as compiling probabilistic programs on-the-fly to one of a suite of computational backends for increased speed. PyMC allows for model specification in Python code, rather than in a domain-specific language, making it easy to learn, customize, and debug. This paper is a tutorial-style introduction to this software package for those already somewhat familiar with Bayesian statistics.
Introduction#
Probabilistic programming (PP) allows flexible specification of Bayesian statistical models in code. PyMC is a PP framework with an intuitive and readable, yet powerful, syntax that is close to the natural syntax statisticians use to describe models. It features next-generation Markov chain Monte Carlo (MCMC) sampling algorithms such as the No-U-Turn Sampler (NUTS; Hoffman, 2014), a self-tuning variant of Hamiltonian Monte Carlo (HMC; Duane, 1987). This class of samplers works well on high-dimensional and complex posterior distributions and allows many complex models to be fit without specialized knowledge about fitting algorithms. HMC and NUTS take advantage of gradient information from the likelihood to achieve much faster convergence than traditional sampling methods, especially for larger models. NUTS also has several self-tuning strategies for adaptively setting the tunable parameters of Hamiltonian Monte Carlo, which means you usually don’t need to have specialized knowledge about how the algorithms work.
Probabilistic programming in Python confers a number of advantages including multi-platform compatibility, an expressive yet clean and readable syntax, easy integration with other scientific libraries, and extensibility via C, C++, Fortran or Cython. These features make it relatively straightforward to write and use custom statistical distributions, samplers and transformation functions, as required by Bayesian analysis.
While most of PyMC’s user-facing features are written in pure Python, it leverages PyTensor (a fork of the Theano project) to transparently transcode models to C and compile them to machine code, thereby boosting performance. PyTensor is a library that allows expressions to be defined using generalized vector data structures called tensors, which are tightly integrated with the popular NumPy ndarray data structure, and similarly allow for broadcasting and advanced indexing, just as NumPy arrays do. PyTensor also automatically optimizes the likelihood’s computational graph for speed and allows for compilation to a suite of computational backends, including Jax and Numba.
Here, we present a primer on the use of PyMC for solving general Bayesian statistical inference and prediction problems. We will first see the basics of how to use PyMC, motivated by a simple example: installation, data creation, model definition, model fitting and posterior analysis. Then we will cover two case studies and use them to show how to define and fit more sophisticated models. Finally we will discuss a couple of other useful features: custom distributions and arbitrary deterministic nodes.
Installation#
Running PyMC requires a relatively recent Python interpreter, preferably version 3.8 or greater. A complete Python installation for macOS, Linux and Windows can most easily be obtained by downloading and installing the free Anaconda Python Distribution by ContinuumIO or the open source Miniforge.
Once Python is installed, follow the installation guide on the PyMC documentation site.
PyMC is distributed under the liberal Apache License 2.0. On the GitHub site, users may also report bugs and other issues, as well as contribute documentation or code to the project, which we actively encourage.
A Motivating Example: Linear Regression#
To introduce model definition, fitting, and posterior analysis, we first consider a simple Bayesian linear regression model with normally-distributed priors for the parameters. We are interested in predicting outcomes \(Y\) as normally-distributed observations with an expected value \(\mu\) that is a linear function of two predictor variables, \(X_1\) and \(X_2\):
where \(\alpha\) is the intercept, and \(\beta_i\) is the coefficient for covariate \(X_i\), while \(\sigma\) represents the observation error. Since we are constructing a Bayesian model, we must assign a prior distribution to the unknown variables in the model. We choose zero-mean normal priors with variance of 100 for both regression coefficients, which corresponds to weak information regarding the true parameter values. We choose a half-normal distribution (normal distribution bounded at zero) as the prior for \(\sigma\).
Generating data#
We can simulate some artificial data from this model using only NumPy’s random module, and then use PyMC to try to recover the corresponding parameters. We are intentionally generating the data to closely correspond the PyMC model structure.
import arviz as az
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
%config InlineBackend.figure_format = 'retina'
# Initialize random number generator
RANDOM_SEED = 8927
rng = np.random.default_rng(RANDOM_SEED)
az.style.use("arviz-darkgrid")
# True parameter values
alpha, sigma = 1, 1
beta = [1, 2.5]
# Size of dataset
size = 100
# Predictor variable
X1 = np.random.randn(size)
X2 = np.random.randn(size) * 0.2
# Simulate outcome variable
Y = alpha + beta[0] * X1 + beta[1] * X2 + rng.normal(size=size) * sigma
Here is what the simulated data look like. We use the pylab module from the plotting library matplotlib.
Model Specification#
Specifying this model in PyMC is straightforward because the syntax is similar to the statistical notation. For the most part, each line of Python code corresponds to a line in the model notation above.
First, we import PyMC. We use the convention of importing it as pm.
import pymc as pm
print(f"Running on PyMC v{pm.__version__}")
Running on PyMC v5.13.1+0.g34c2d31.dirty
Now we build our model, which we will present in full first, then explain each part line-by-line.
basic_model = pm.Model()
with basic_model:
# Priors for unknown model parameters
alpha = pm.Normal("alpha", mu=0, sigma=10)
beta = pm.Normal("beta", mu=0, sigma=10, shape=2)
sigma = pm.HalfNormal("sigma", sigma=1)
# Expected value of outcome
mu = alpha + beta[0] * X1 + beta[1] * X2
# Likelihood (sampling distribution) of observations
Y_obs = pm.Normal("Y_obs", mu=mu, sigma=sigma, observed=Y)
The first line,
basic_model = pm.Model()
creates a new Model object which is a container for the model random variables.
Following instantiation of the model, the subsequent specification of the model components is performed inside a with statement:
with basic_model:
This creates a context manager, with our basic_model as the context, that includes all statements until the indented block ends. This means all PyMC objects introduced in the indented code block below the with statement are added to the model behind the scenes. Absent this context manager idiom, we would be forced to manually associate each of the variables with basic_model right after we create them. If you try to create a new random variable without a with model: statement, it will raise an error since there is no obvious model for the variable to be added to.
The first three statements in the context manager:
alpha = pm.Normal('alpha', mu=0, sigma=10)
beta = pm.Normal('beta', mu=0, sigma=10, shape=2)
sigma = pm.HalfNormal('sigma', sigma=1)
create stochastic random variables with normally-distributed prior distributions for the regression coefficients with a mean of 0 and standard deviation of 10, and a half-normal distribution for the standard deviation of the observations, \(\sigma\). These are stochastic because their values are partly determined by their parents in the dependency graph of random variables, which for priors are simple constants, and partly random (or stochastic).
We call the pm.Normal constructor to create a random variable to use as a normal prior. The first argument is always the name of the random variable, which should almost always match the name of the Python variable being assigned to, since it is sometimes used to retrieve the variable from the model for summarizing output. The remaining required arguments for a stochastic object are the parameters, in this case mu, the mean, and sigma, the standard deviation, which we assign hyperparameter values for the model. In general, a distribution’s parameters are values that determine the location, shape or scale of the random variable, depending on the parameterization of the distribution. Most commonly-used distributions, such as Beta, Exponential, Categorical, Gamma, Binomial and many others, are available in PyMC.
The beta variable has an additional shape argument to denote it as a vector-valued parameter of size 2. The shape argument is available for all distributions and specifies the length or shape of the random variable, but is optional for scalar variables, since it defaults to a value of one. It can be an integer to specify an array, or a tuple to specify a multidimensional array (e.g. shape=(5,7) makes a random variable that takes on 5 by 7 matrix values).
Detailed notes about distributions, sampling methods and other PyMC functions are available in the API documentation.
Having defined the priors, the next statement creates the expected value mu of the outcomes, specifying the linear relationship:
mu = alpha + beta[0]*X1 + beta[1]*X2
This creates a deterministic random variable, which implies that its value is completely determined by its parents’ values. That is, there is no uncertainty beyond that which is inherent in the parents’ values. Here, mu is just the sum of the intercept alpha and the two products of the coefficients in beta and the predictor variables, whatever their values may be.
PyMC random variables and data can be arbitrarily added, subtracted, divided, multiplied together and indexed-into to create new random variables. This allows for great model expressivity. Many common mathematical functions like sum, sin, exp and linear algebra functions like dot (for inner product) and inv (for inverse) are also provided.
The final line of the model defines Y_obs, the sampling distribution of the outcomes in the dataset.
Y_obs = Normal('Y_obs', mu=mu, sigma=sigma, observed=Y)
This is a special case of a stochastic variable that we call an observed stochastic, and represents the data likelihood of the model. It is identical to a standard stochastic, except that its observed argument, which passes the data to the variable, indicates that the values for this variable were observed, and should not be changed by any fitting algorithm applied to the model. The data can be passed in the form of either a ndarray or DataFrame object.
Notice that, unlike for the priors of the model, the parameters for the normal distribution of Y_obs are not fixed values, but rather are the deterministic object mu and the stochastic sigma. This creates parent-child relationships between the likelihood and these two variables.
with basic_model:
# draw 1000 posterior samples
idata = pm.sample()
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (2 chains in 1 job)
NUTS: [alpha, beta, sigma]
Sampling 2 chains for 1_000 tune and 1_000 draw iterations (2_000 + 2_000 draws total) took 4 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
The sample function runs the step method(s) assigned (or passed) to it for the given number of iterations and returns an InferenceData object containing the samples collected, along with other useful attributes like statistics of the sampling run and a copy of the observed data. Notice that sample generated a set of parallel chains, depending on how many compute cores are on your machine.
idata
-
<xarray.Dataset> Size: 72kB Dimensions: (chain: 2, draw: 1000, beta_dim_0: 2) Coordinates: * chain (chain) int64 16B 0 1 * draw (draw) int64 8kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * beta_dim_0 (beta_dim_0) int64 16B 0 1 Data variables: alpha (chain, draw) float64 16kB 1.313 0.9756 1.251 ... 1.234 1.225 beta (chain, draw, beta_dim_0) float64 32kB 0.9983 4.017 ... 3.941 sigma (chain, draw) float64 16kB 0.9002 0.9234 1.105 ... 0.8263 0.8738 Attributes: created_at: 2024-04-15T09:24:21.011577+00:00 arviz_version: 0.18.0 inference_library: pymc inference_library_version: 5.13.1+0.g34c2d31.dirty sampling_time: 3.7472190856933594 tuning_steps: 1000 -
<xarray.Dataset> Size: 252kB Dimensions: (chain: 2, draw: 1000) Coordinates: * chain (chain) int64 16B 0 1 * draw (draw) int64 8kB 0 1 2 3 4 5 ... 995 996 997 998 999 Data variables: (12/17) acceptance_rate (chain, draw) float64 16kB 0.7436 0.8502 ... 1.0 1.0 diverging (chain, draw) bool 2kB False False ... False False energy (chain, draw) float64 16kB 149.9 151.0 ... 148.7 energy_error (chain, draw) float64 16kB 0.05631 ... -0.4417 index_in_trajectory (chain, draw) int64 16kB -1 -2 -2 -2 -1 ... 2 -2 3 1 largest_eigval (chain, draw) float64 16kB nan nan nan ... nan nan ... ... process_time_diff (chain, draw) float64 16kB 0.0005767 ... 0.0002827 reached_max_treedepth (chain, draw) bool 2kB False False ... False False smallest_eigval (chain, draw) float64 16kB nan nan nan ... nan nan step_size (chain, draw) float64 16kB 1.04 1.04 ... 0.9807 step_size_bar (chain, draw) float64 16kB 1.007 1.007 ... 0.9801 tree_depth (chain, draw) int64 16kB 2 2 2 2 1 2 ... 2 3 2 2 2 1 Attributes: created_at: 2024-04-15T09:24:21.024254+00:00 arviz_version: 0.18.0 inference_library: pymc inference_library_version: 5.13.1+0.g34c2d31.dirty sampling_time: 3.7472190856933594 tuning_steps: 1000 -
<xarray.Dataset> Size: 2kB Dimensions: (Y_obs_dim_0: 100) Coordinates: * Y_obs_dim_0 (Y_obs_dim_0) int64 800B 0 1 2 3 4 5 6 ... 93 94 95 96 97 98 99 Data variables: Y_obs (Y_obs_dim_0) float64 800B 0.4931 2.704 ... -0.007793 2.713 Attributes: created_at: 2024-04-15T09:24:21.029679+00:00 arviz_version: 0.18.0 inference_library: pymc inference_library_version: 5.13.1+0.g34c2d31.dirty
The various attributes of the InferenceData object can be queried in a similar way to a dict containing a map from variable names to numpy.arrays. For example, we can retrieve the sampling trace from the alpha latent variable by using the variable name as an index to the idata.posterior attribute. The first dimension of the returned array is the chain index, the second dimension is the sampling index, while the later dimensions match the shape of the variable. We can see the first 5 values for the alpha variable in each chain as follows:
idata.posterior["alpha"].sel(draw=slice(0, 4))
<xarray.DataArray 'alpha' (chain: 2, draw: 5)> Size: 80B
array([[1.31264777, 0.97564685, 1.25091896, 1.20025368, 1.19146959],
[1.09672998, 1.09431182, 1.10105174, 1.24687676, 1.10451708]])
Coordinates:
* chain (chain) int64 16B 0 1
* draw (draw) int64 40B 0 1 2 3 4If we wanted to use the slice sampling algorithm to sample our parameters instead of NUTS (which was assigned automatically), we could have specified this as the step argument for sample.
with basic_model:
# instantiate sampler
step = pm.Slice()
# draw 5000 posterior samples
slice_idata = pm.sample(5000, step=step)
Sequential sampling (2 chains in 1 job)
CompoundStep
>Slice: [alpha]
>Slice: [beta]
>Slice: [sigma]
Sampling 2 chains for 1_000 tune and 5_000 draw iterations (2_000 + 10_000 draws total) took 8 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
Posterior analysis#
PyMC’s plotting and diagnostics functionalities are now taken care of by a dedicated, platform-agnostic package named Arviz. A simple posterior plot can be created using plot_trace.
The left column consists of a smoothed histogram (using kernel density estimation) of the marginal posteriors of each stochastic random variable while the right column contains the samples of the Markov chain plotted in sequential order. The beta variable, being vector-valued, produces two density plots and two trace plots, corresponding to both predictor coefficients.
In addition, the summary function provides a text-based output of common posterior statistics:
az.summary(idata, round_to=2)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 1.17 | 0.10 | 0.98 | 1.35 | 0.00 | 0.00 | 2553.01 | 1493.16 | 1.0 |
| beta[0] | 0.90 | 0.10 | 0.69 | 1.09 | 0.00 | 0.00 | 3107.57 | 1590.29 | 1.0 |
| beta[1] | 3.90 | 0.44 | 3.07 | 4.70 | 0.01 | 0.01 | 3512.09 | 1258.82 | 1.0 |
| sigma | 0.96 | 0.07 | 0.83 | 1.09 | 0.00 | 0.00 | 3211.75 | 1577.55 | 1.0 |
Case study 1: Educational Outcomes for Hearing-impaired Children#
As a motivating example, we will use a dataset of educational outcomes for children with hearing impairment. Here, we are interested in determining factors that are associated with better or poorer learning outcomes.
The Data#
This anonymized dataset is taken from the Listening and Spoken Language Data Repository (LSL-DR), an international data repository that tracks the demographics and longitudinal outcomes for children who have hearing loss and are enrolled in programs focused on supporting listening and spoken language development. Researchers are interested in discovering factors related to improvements in educational outcomes within these programs.
There is a suite of available predictors, including:
gender (
male)number of siblings in the household (
siblings)index of family involvement (
family_inv)whether the primary household language is not English (
non_english)presence of a previous disability (
prev_disab)non-white race (
non_white)age at the time of testing (in months,
age_test)whether hearing loss is not severe (
non_severe_hl)whether the subject’s mother obtained a high school diploma or better (
mother_hs)whether the hearing impairment was identified by 3 months of age (
early_ident).
The outcome variable is a standardized test score in one of several learning domains.
test_scores = pd.read_csv(pm.get_data("test_scores.csv"), index_col=0)
test_scores.head()
| score | male | siblings | family_inv | non_english | prev_disab | age_test | non_severe_hl | mother_hs | early_ident | non_white | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 40 | 0 | 2.0 | 2.0 | False | NaN | 55 | 1.0 | NaN | False | False |
| 1 | 31 | 1 | 0.0 | NaN | False | 0.0 | 53 | 0.0 | 0.0 | False | False |
| 2 | 83 | 1 | 1.0 | 1.0 | True | 0.0 | 52 | 1.0 | NaN | False | True |
| 3 | 75 | 0 | 3.0 | NaN | False | 0.0 | 55 | 0.0 | 1.0 | False | False |
| 5 | 62 | 0 | 0.0 | 4.0 | False | 1.0 | 50 | 0.0 | NaN | False | False |
# Dropping missing values is a very bad idea in general, but we do so here for simplicity
X = test_scores.dropna().astype(float)
y = X.pop("score")
# Standardize the features
X -= X.mean()
X /= X.std()
N, D = X.shape
The Model#
This is a more realistic problem than the first regression example, as we are now dealing with a multivariate regression model. However, while there are several potential predictors in the LSL-DR dataset, it is difficult a priori to determine which ones are relevant for constructing an effective statistical model. There are a number of approaches for conducting variable selection, but a popular automated method is regularization, whereby ineffective covariates are shrunk towards zero via regularization (a form of penalization) if they do not contribute to predicting outcomes.
You may have heard of regularization from machine learning or classical statistics applications, where methods like the lasso or ridge regression shrink parameters towards zero by applying a penalty to the size of the regression parameters. In a Bayesian context, we apply an appropriate prior distribution to the regression coefficients. One such prior is the hierarchical regularized horseshoe, which uses two regularization strategies, one global and a set of local parameters, one for each coefficient. The key to making this work is by selecting a long-tailed distribution as the shrinkage priors, which allows some to be nonzero, while pushing the rest towards zero.
The horeshoe prior for each regression coefficient \(\beta_i\) looks like this:
where \(\sigma\) is the prior on the error standard deviation that will also be used for the model likelihood. Here, \(\tau\) is the global shrinkage parameter and \(\tilde{\lambda}_i\) is the local shrinkage parameter. Let’s start global: for the prior on \(\tau\) we will use a Half-StudentT distribution, which is a reasonable choice becuase it is heavy-tailed.
One catch is that the parameterization of the prior requires a pre-specified value \(D_0\), which represents the true number of non-zero coefficients. Fortunately, a reasonable guess at this value is all that is required, and it need only be within an order of magnitude of the true number. Let’s use half the number of predictors as our guess:
D0 = int(D / 2)
Meanwhile, the local shrinkage parameters are defined by the ratio:
To complete this specification, we need priors on \(\lambda_i\) and \(c\); as with the global shrinkage, we use a long-tailed \(\textrm{Half-StudentT}_5(1)\) on the \(\lambda_i\). We need \(c^2\) to be strictly positive, but not necessarily long-tailed, so an inverse gamma prior on \(c^2\), \(c^2 \sim \textrm{InverseGamma}(1, 1)\) fits the bill.
Finally, to allow the NUTS sampler to sample the \(\beta_i\) more efficiently, we will re-parameterize it as follows:
You will run into this reparameterization a lot in practice.
Model Specification#
Specifying the model in PyMC mirrors its statistical specification. This model employs a couple of new distributions: the HalfStudentT distribution for the \(\tau\) and \(\lambda\) priors, and the InverseGamma distribution for the \(c^2\) variable.
In PyMC, variables with purely positive priors like InverseGamma are transformed with a log transform. This makes sampling more robust. Behind the scenes, a variable in the unconstrained space (named <variable-name>_log) is added to the model for sampling. Variables with priors that constrain them on two sides, like Beta or Uniform, are also transformed to be unconstrained but with a log odds transform.
We are also going to take advantage of named dimensions in PyMC and ArviZ by passing the input variable names into the model as coordinates called “predictors”. This will allow us to pass this vector of names as a replacement for the shape integer argument in the vector-valued parameters. The model will then associate the appropriate name with each latent parameter that it is estimating. This is a little more work to set up, but will pay dividends later when we are working with our model output.
Let’s encode this model in PyMC:
import pytensor.tensor as at
with pm.Model(coords={"predictors": X.columns.values}) as test_score_model:
# Prior on error SD
sigma = pm.HalfNormal("sigma", 25)
# Global shrinkage prior
tau = pm.HalfStudentT("tau", 2, D0 / (D - D0) * sigma / np.sqrt(N))
# Local shrinkage prior
lam = pm.HalfStudentT("lam", 5, dims="predictors")
c2 = pm.InverseGamma("c2", 1, 1)
z = pm.Normal("z", 0.0, 1.0, dims="predictors")
# Shrunken coefficients
beta = pm.Deterministic(
"beta", z * tau * lam * at.sqrt(c2 / (c2 + tau**2 * lam**2)), dims="predictors"
)
# No shrinkage on intercept
beta0 = pm.Normal("beta0", 100, 25.0)
scores = pm.Normal("scores", beta0 + at.dot(X.values, beta), sigma, observed=y.values)
Notice that we have wrapped the calculation of beta in a Deterministic PyMC class. You can read more about this in detail below, but this ensures that the values of this deterministic variable is retained in the sample trace.
Also note that we have declared the Model name test_score_model in the first occurrence of the context manager, rather than splitting it into two lines, as we did for the first example.
Once the model is complete, we can look at its structure using GraphViz, which plots the model graph. It’s useful to ensure that the relationships in the model you have coded are correct, as it’s easy to make coding mistakes.
Before we proceed further, let’s see what the model does before it sees any data. We can conduct prior predictive sampling to generate simulated data from the model. Then, let’s compare these simulations to the actual test scores in the dataset.
with test_score_model:
prior_samples = pm.sample_prior_predictive(100)
Sampling: [beta0, c2, lam, scores, sigma, tau, z]
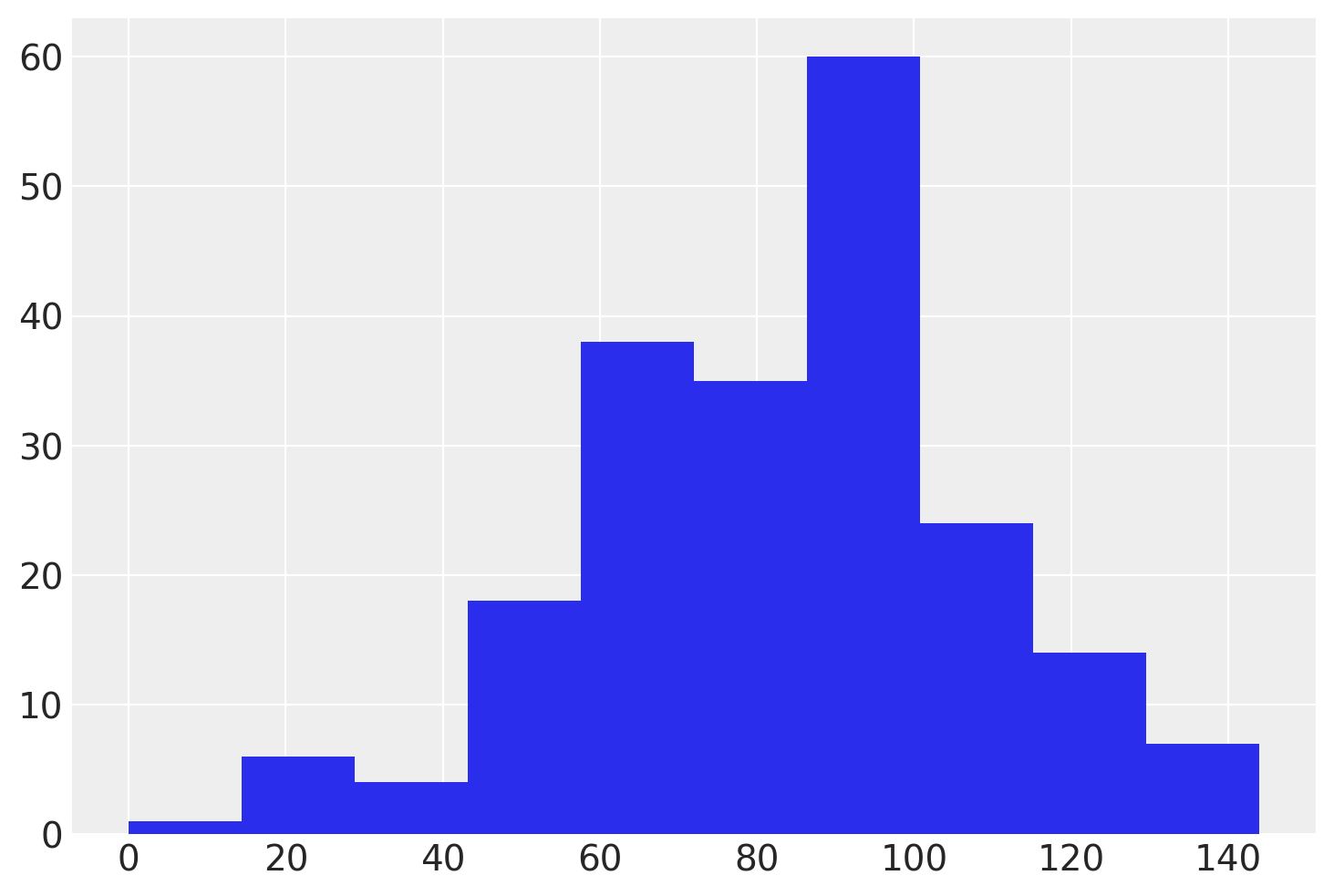
az.plot_dist(
test_scores["score"].values,
kind="hist",
color="C1",
hist_kwargs=dict(alpha=0.6),
label="observed",
)
az.plot_dist(
prior_samples.prior_predictive["scores"],
kind="hist",
hist_kwargs=dict(alpha=0.6),
label="simulated",
)
plt.xticks(rotation=45);
How do we know if this is reasonable or not? This requires some domain knowledge of the problem. Here, we are trying to predict the outcomes of test scores. If our model was predicting values in the thousands, or lots of negative values, while excluding scores that are plausible, then we have misspecified our model. You can see here that the support of the distribution of simulated data completely overlaps the support of the observed distribution of scores; this is a good sign! There are a few negative values and a few that are probably too large to be plausible, but nothing to worry about.
Model Fitting#
Now for the easy part: PyMC’s “Inference Button” is the call to sample. We will let this model tune for a little longer than the default value (1000 iterations). This gives the NUTS sampler a little more time to tune itself adequately.
with test_score_model:
idata = pm.sample(1000, tune=2000, random_seed=42)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (2 chains in 1 job)
NUTS: [sigma, tau, lam, c2, z, beta0]
Sampling 2 chains for 2_000 tune and 1_000 draw iterations (4_000 + 2_000 draws total) took 31 seconds.
There were 2 divergences after tuning. Increase `target_accept` or reparameterize.
We recommend running at least 4 chains for robust computation of convergence diagnostics
Notice that we have a few warnings here about divergences. These are samples where NUTS was not able to make a valid move across the posterior distribution, so the resulting points are probably not representative samples from the posterior. There aren’t many in this example, so it’s nothing to worry about, but let’s go ahead and follow the advice and increase target_accept from its default value of 0.9 to 0.99.
with test_score_model:
idata = pm.sample(1000, tune=2000, random_seed=42, target_accept=0.99)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (2 chains in 1 job)
NUTS: [sigma, tau, lam, c2, z, beta0]
Sampling 2 chains for 2_000 tune and 1_000 draw iterations (4_000 + 2_000 draws total) took 102 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
Since the target acceptance rate is larger, the algorithm is being more conservative with its leapfrog steps, making them smaller. The price we pay for this is that each sample takes longer to complete. However, the warnings are now gone, and we have a clean posterior sample!
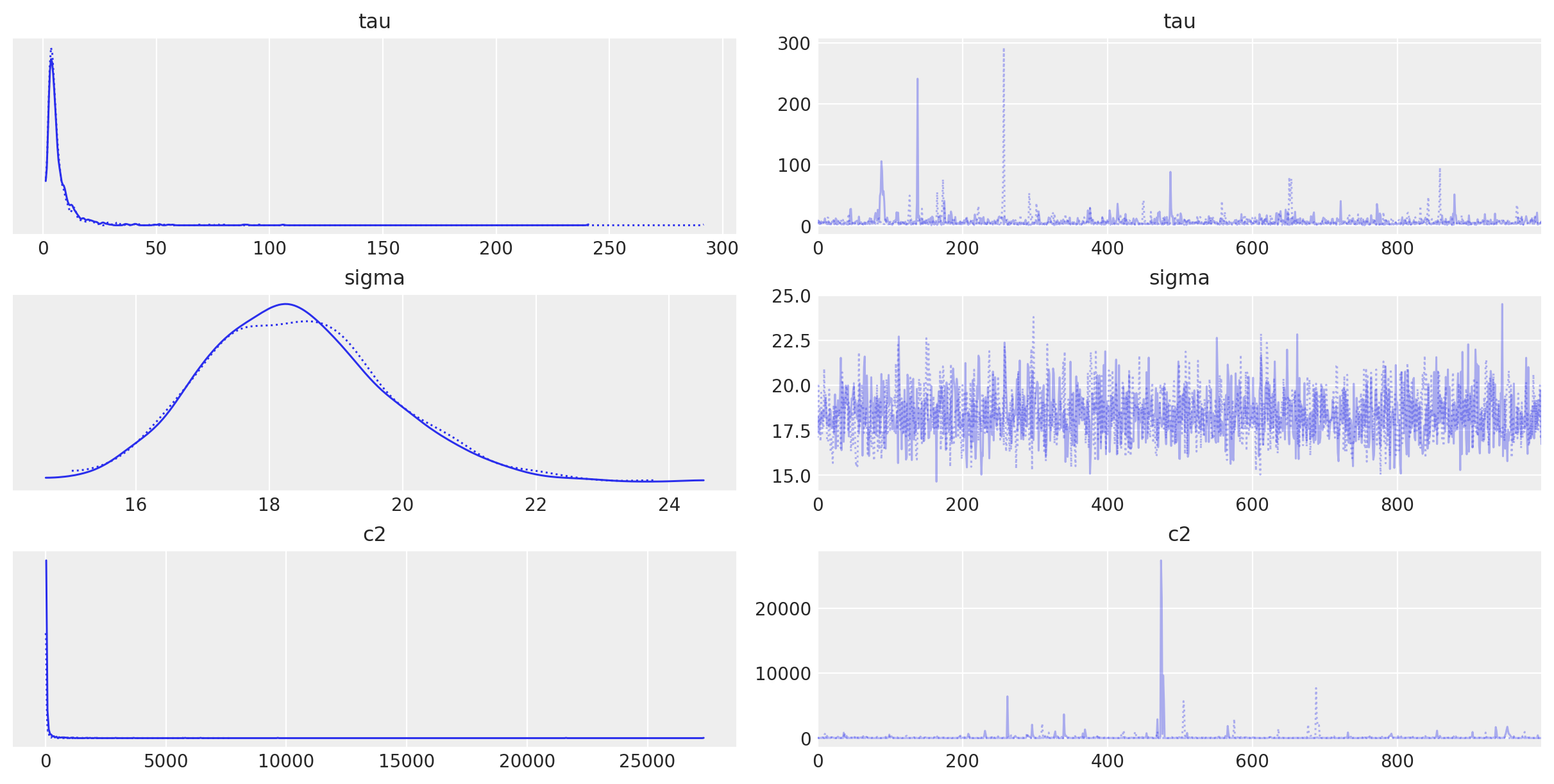
Model Checking#
A simple first step in model checking is to visually inspect our samples by looking at the traceplot for the univariate latent parameters to check for obvious problems. These names can be passed to plot_trace in the var_names argument.
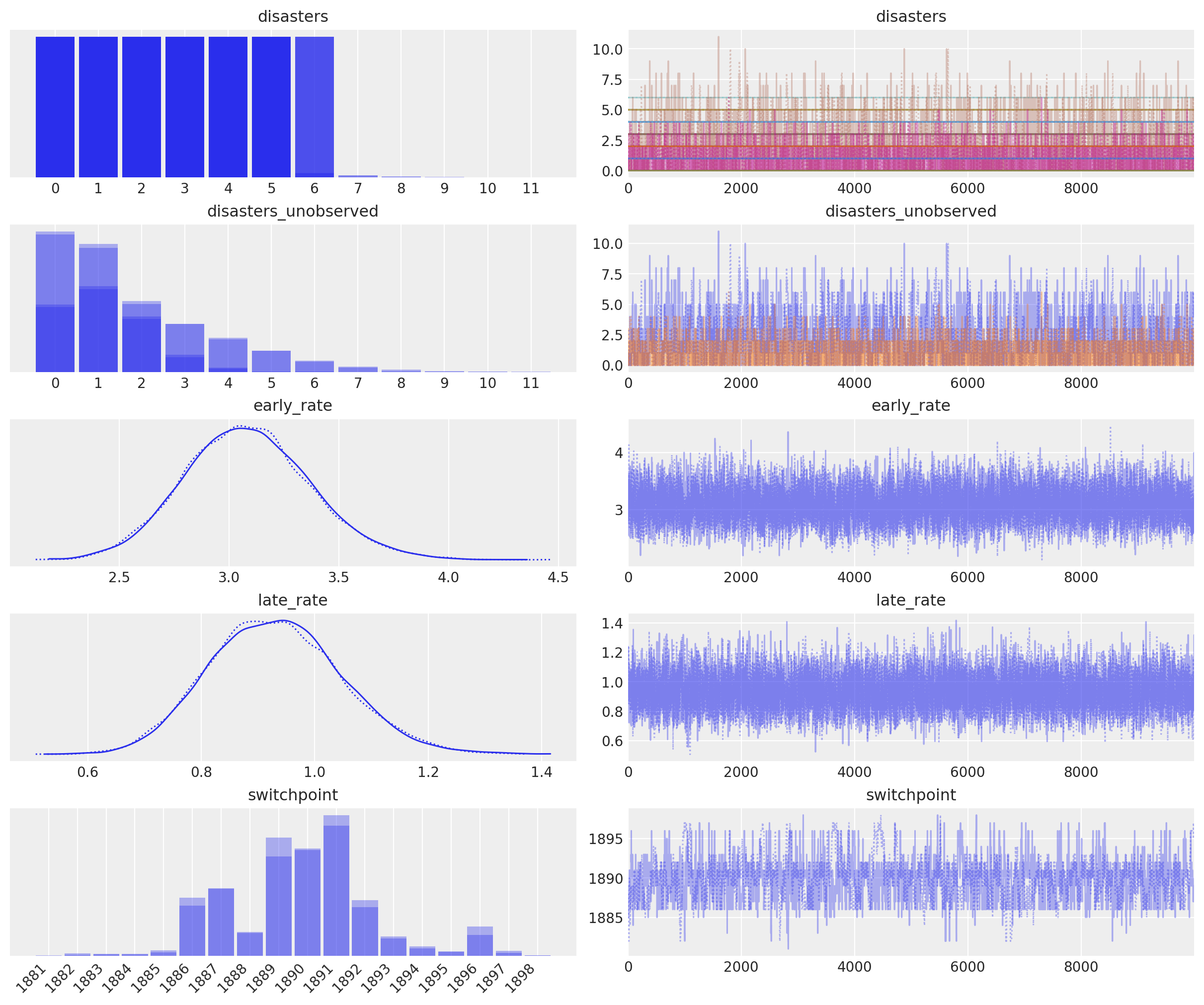
Do these look okay? Well, each of the densities on the left side for each parameter look pretty similar to the others, which means they have converged to the same posterior distribution (be it the correct one or not). The homogeneity of the trace plots on the right are also a good sign; there is no trend or pattern to the time series of sampled values. Note that c2 and tau occasionally sample extreme values, but this is expected from heavy-tailed distributions.
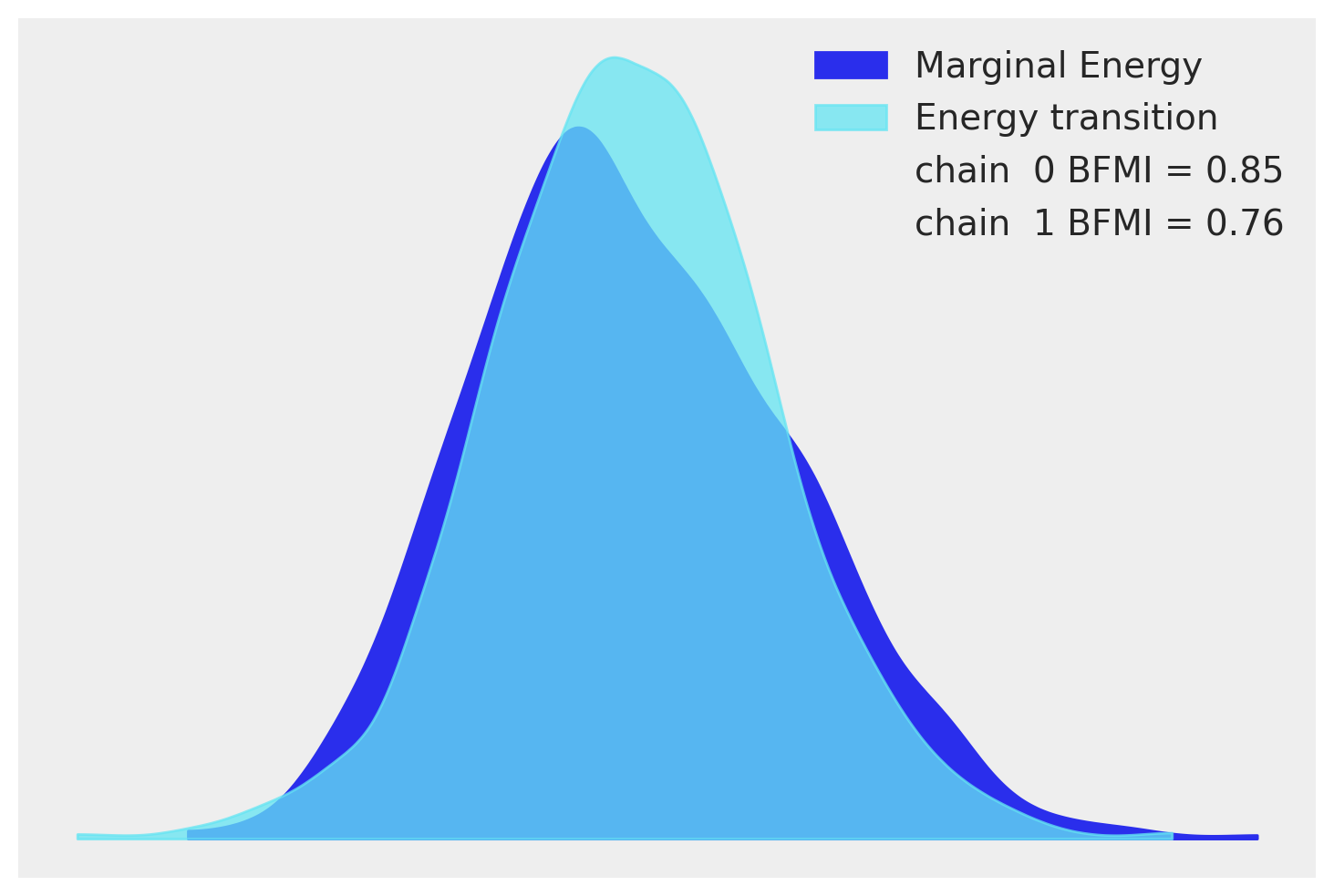
The next easy model-checking step is to see if the NUTS sampler performed as expected. An energy plot is a way of checking if the NUTS algorithm was able to adequately explore the posterior distribution. If it was not, one runs the risk of biased posterior estimates when parts of the posterior are not visited with adequate frequency. The plot shows two density estimates: one is the marginal energy distribution of the sampling run and the other is the distribution of the energy transitions between steps. This is all a little abstract, but all we are looking for is for the distributions to be similar to one another. Ours does not look too bad.
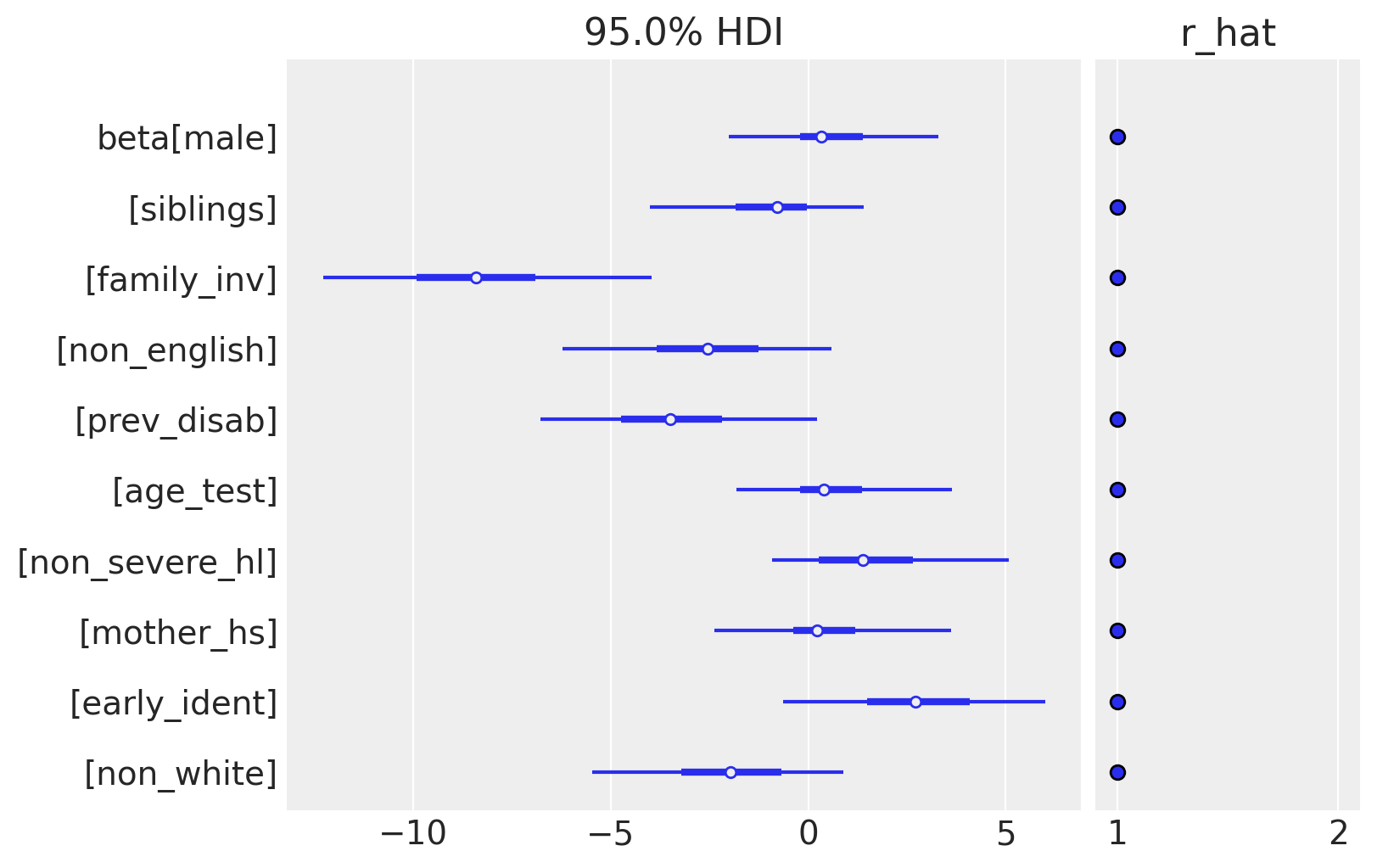
Ultimately, we are interested in the estimates of beta, the set of predictor coefficients. Passing beta to plot_trace would generate a very crowded plot, so we will use plot_forest instead, which is designed to handle vector-valued parameters.
The posterior distribution of coefficients reveal some factors that appear to be important in predicting test scores. Family involvement (family_inv) is large and negative, meaning a larger score (which is related to poorer involvement) results in much worse test scores. On the other end, early identification of hearing impairment is positive, meaning that detecting a problem early results in better educational outcomes down the road, which is also intuitive. Notice that other variables, notably gender (male), age at testing (age_test), and the mother’s educational status (mother_hs) have all been shrunk essentially to zero.
Case study 2: Coal mining disasters#
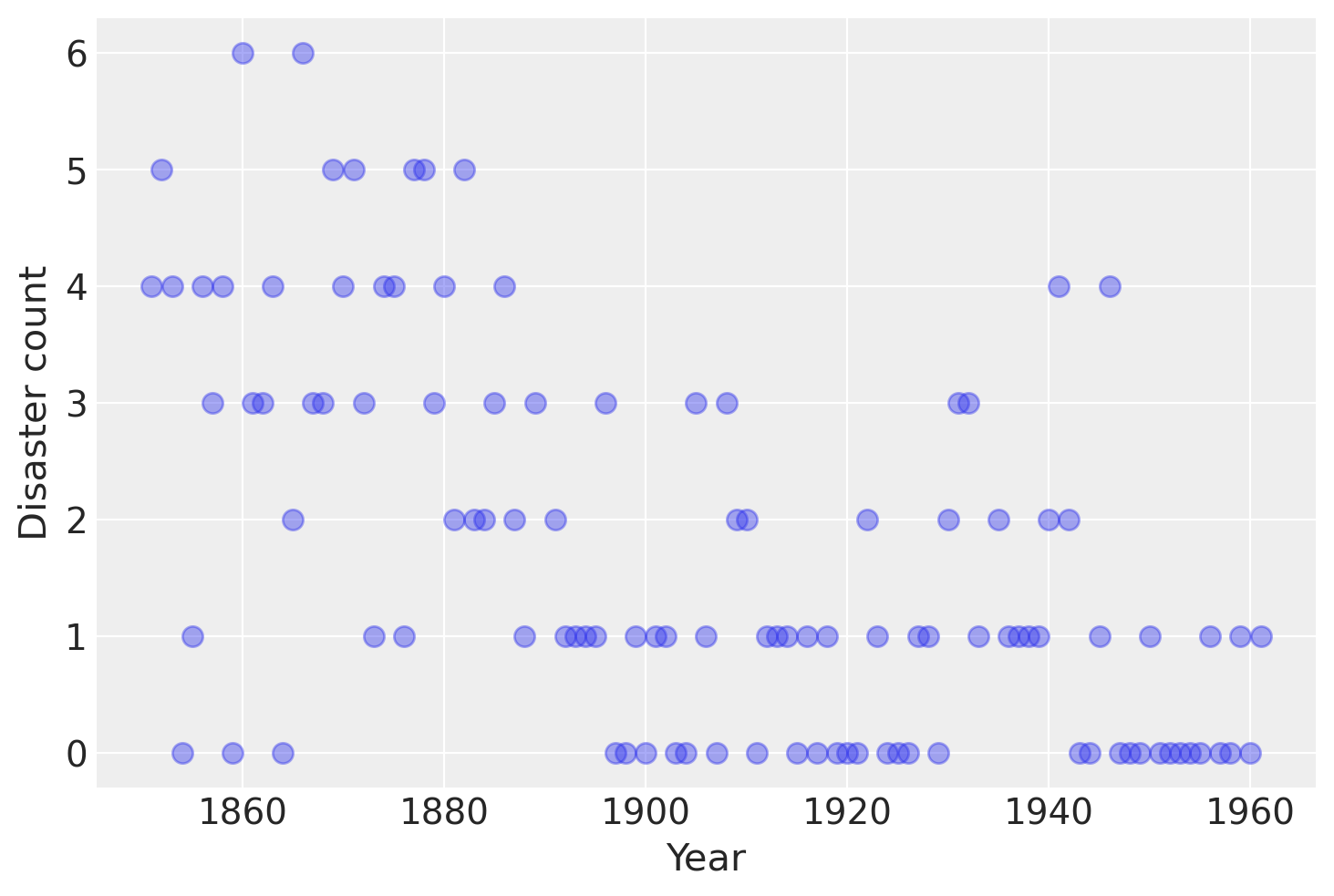
Consider the following time series of recorded coal mining disasters in the UK from 1851 to 1962 (Jarrett, 1979). The number of disasters is thought to have been affected by changes in safety regulations during this period. Unfortunately, we also have a pair of years with missing data, identified as missing by a nan in the pandas Series. These missing values will be automatically imputed by PyMC.
Next we will build a model for this series and attempt to estimate when the change occurred. At the same time, we will see how to handle missing data, use multiple samplers and sample from discrete random variables.
# fmt: off
disaster_data = pd.Series(
[4, 5, 4, 0, 1, 4, 3, 4, 0, 6, 3, 3, 4, 0, 2, 6,
3, 3, 5, 4, 5, 3, 1, 4, 4, 1, 5, 5, 3, 4, 2, 5,
2, 2, 3, 4, 2, 1, 3, np.nan, 2, 1, 1, 1, 1, 3, 0, 0,
1, 0, 1, 1, 0, 0, 3, 1, 0, 3, 2, 2, 0, 1, 1, 1,
0, 1, 0, 1, 0, 0, 0, 2, 1, 0, 0, 0, 1, 1, 0, 2,
3, 3, 1, np.nan, 2, 1, 1, 1, 1, 2, 4, 2, 0, 0, 1, 4,
0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1]
)
# fmt: on
years = np.arange(1851, 1962)
plt.plot(years, disaster_data, "o", markersize=8, alpha=0.4)
plt.ylabel("Disaster count")
plt.xlabel("Year");
Occurrences of disasters in the time series is thought to follow a Poisson process with a large rate parameter in the early part of the time series, and from one with a smaller rate in the later part. We are interested in locating the change point in the series, which is perhaps related to changes in mining safety regulations.
In our model,
the parameters are defined as follows:
\(D_t\): The number of disasters in year \(t\)
\(r_t\): The rate parameter of the Poisson distribution of disasters in year \(t\).
\(s\): The year in which the rate parameter changes (the switchpoint).
\(e\): The rate parameter before the switchpoint \(s\).
\(l\): The rate parameter after the switchpoint \(s\).
\(t_l\), \(t_h\): The lower and upper boundaries of year \(t\).
This model is built much like our previous models. The major differences are the introduction of discrete variables with the Poisson and discrete-uniform priors and the novel form of the deterministic random variable rate.
with pm.Model() as disaster_model:
switchpoint = pm.DiscreteUniform("switchpoint", lower=years.min(), upper=years.max())
# Priors for pre- and post-switch rates number of disasters
early_rate = pm.Exponential("early_rate", 1.0)
late_rate = pm.Exponential("late_rate", 1.0)
# Allocate appropriate Poisson rates to years before and after current
rate = pm.math.switch(switchpoint >= years, early_rate, late_rate)
disasters = pm.Poisson("disasters", rate, observed=disaster_data)
/home/docs/checkouts/readthedocs.org/user_builds/pymc/conda/stable/lib/python3.11/site-packages/pymc/model/core.py:1312: RuntimeWarning: invalid value encountered in cast
data = convert_observed_data(data).astype(rv_var.dtype)
/home/docs/checkouts/readthedocs.org/user_builds/pymc/conda/stable/lib/python3.11/site-packages/pymc/model/core.py:1326: ImputationWarning: Data in disasters contains missing values and will be automatically imputed from the sampling distribution.
warnings.warn(impute_message, ImputationWarning)
The logic for the rate random variable,
rate = switch(switchpoint >= year, early_rate, late_rate)
is implemented using switch, a function that works like an if statement. It uses the first argument to switch between the next two arguments.
Missing values are handled transparently by passing a NumPy MaskedArray or a DataFrame with NaN values to the observed argument when creating an observed stochastic random variable. Behind the scenes, another random variable, disasters.missing_values is created to model the missing values.
Unfortunately, because they are discrete variables and thus have no meaningful gradient, we cannot use NUTS for sampling switchpoint or the missing disaster observations. Instead, we will sample using a Metropolis step method, which implements adaptive Metropolis-Hastings, because it is designed to handle discrete values. PyMC automatically assigns the correct sampling algorithms.
with disaster_model:
idata = pm.sample(10000)
Sequential sampling (2 chains in 1 job)
CompoundStep
>CompoundStep
>>Metropolis: [switchpoint]
>>Metropolis: [disasters_unobserved]
>NUTS: [early_rate, late_rate]
Sampling 2 chains for 1_000 tune and 10_000 draw iterations (2_000 + 20_000 draws total) took 31 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
In the trace plot below we can see that there’s about a 10 year span that’s plausible for a significant change in safety, but a 5 year span that contains most of the probability mass. The distribution is jagged because of the jumpy relationship between the year switchpoint and the likelihood; the jaggedness is not due to sampling error.
axes_arr = az.plot_trace(idata)
plt.draw()
for ax in axes_arr.flatten():
if ax.get_title() == "switchpoint":
labels = [label.get_text() for label in ax.get_xticklabels()]
ax.set_xticklabels(labels, rotation=45, ha="right")
break
plt.draw()
Note that the rate random variable does not appear in the trace. That is fine in this case, because it is not of interest in itself. Remember from the previous example, we would trace the variable by wrapping it in a Deterministic class, and giving it a name.
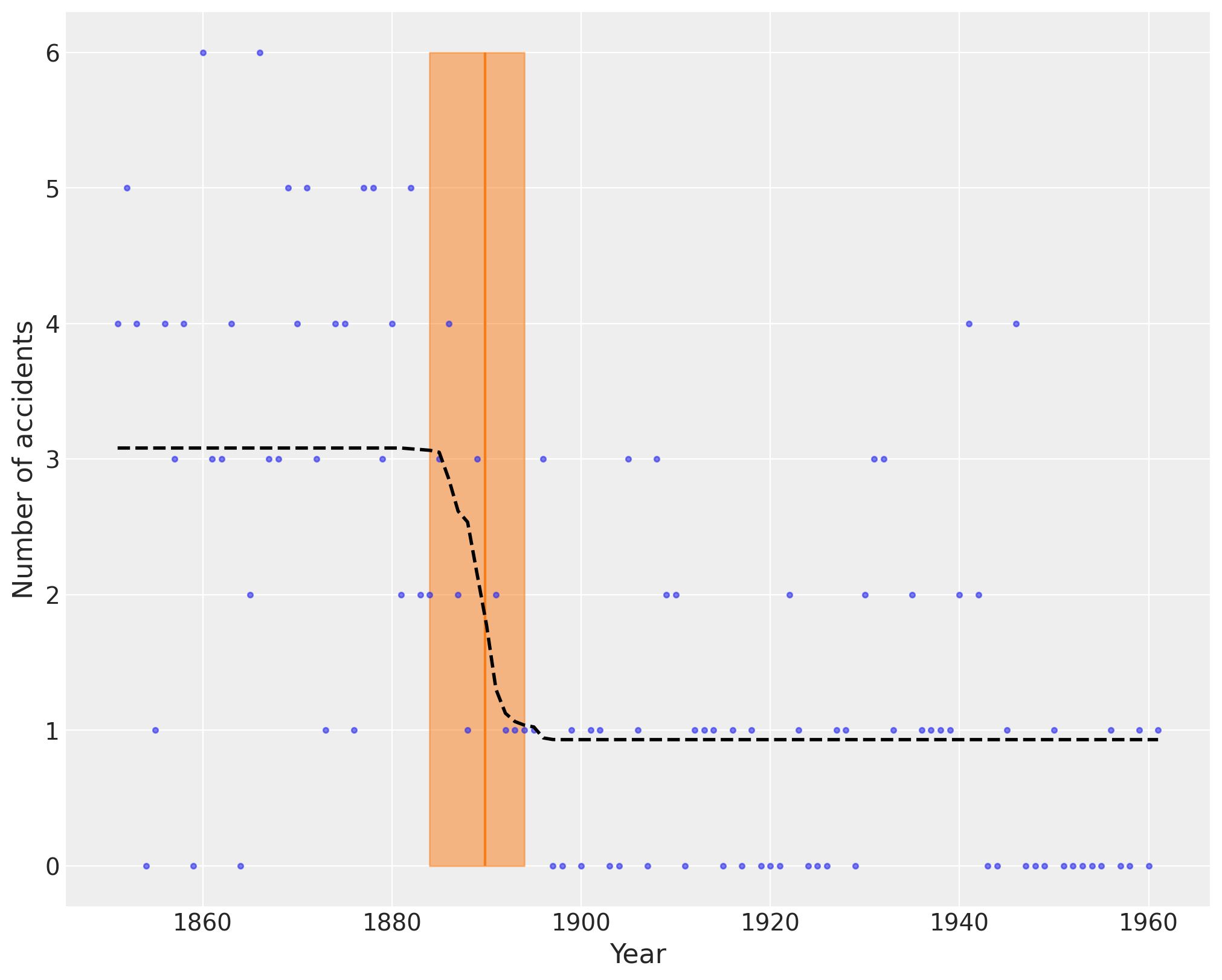
The following plot shows the switch point as an orange vertical line, together with its highest posterior density (HPD) as a semitransparent band. The dashed black line shows the accident rate.
plt.figure(figsize=(10, 8))
plt.plot(years, disaster_data, ".", alpha=0.6)
plt.ylabel("Number of accidents", fontsize=16)
plt.xlabel("Year", fontsize=16)
trace = idata.posterior.stack(draws=("chain", "draw"))
plt.vlines(trace["switchpoint"].mean(), disaster_data.min(), disaster_data.max(), color="C1")
average_disasters = np.zeros_like(disaster_data, dtype="float")
for i, year in enumerate(years):
idx = year < trace["switchpoint"]
average_disasters[i] = np.mean(np.where(idx, trace["early_rate"], trace["late_rate"]))
sp_hpd = az.hdi(idata, var_names=["switchpoint"])["switchpoint"].values
plt.fill_betweenx(
y=[disaster_data.min(), disaster_data.max()],
x1=sp_hpd[0],
x2=sp_hpd[1],
alpha=0.5,
color="C1",
)
plt.plot(years, average_disasters, "k--", lw=2);
Arbitrary deterministics#
Due to its reliance on PyTensor, PyMC provides many mathematical functions and operators for transforming random variables into new random variables. However, the library of functions in PyTensor is not exhaustive, therefore PyTensor and PyMC provide functionality for creating arbitrary functions in pure Python, and including these functions in PyMC models. This is supported with the as_op function decorator.
PyTensor needs to know the types of the inputs and outputs of a function, which are specified for as_op by itypes for inputs and otypes for outputs.
from pytensor.compile.ops import as_op
@as_op(itypes=[at.lscalar], otypes=[at.lscalar])
def crazy_modulo3(value):
if value > 0:
return value % 3
else:
return (-value + 1) % 3
with pm.Model() as model_deterministic:
a = pm.Poisson("a", 1)
b = crazy_modulo3(a)
An important drawback of this approach is that it is not possible for pytensor to inspect these functions in order to compute the gradient required for the Hamiltonian-based samplers. Therefore, it is not possible to use the HMC or NUTS samplers for a model that uses such an operator. However, it is possible to add a gradient if we inherit from Op instead of using as_op. The PyMC example set includes a more elaborate example of the usage of as_op.
Arbitrary distributions#
Similarly, the library of statistical distributions in PyMC is not exhaustive, but PyMC allows for the creation of user-defined functions for an arbitrary probability distribution. For simple statistical distributions, the DensityDist class takes as an argument any function that calculates a log-probability \(log(p(x))\). This function may employ other random variables in its calculation. Here is an example inspired by a blog post by Jake Vanderplas on which priors to use for a linear regression (Vanderplas, 2014).
import pytensor.tensor as at
with pm.Model() as model:
alpha = pm.Uniform('intercept', -100, 100)
# Create custom densities
beta = pm.DensityDist('beta', logp=lambda value: -1.5 * at.log(1 + value**2))
eps = pm.DensityDist('eps', logp=lambda value: -at.log(at.abs_(value)))
# Create likelihood
like = pm.Normal('y_est', mu=alpha + beta * X, sigma=eps, observed=Y)
For more complex distributions, one can create a subclass of Continuous or Discrete and provide the custom logp function, as required. This is how the built-in distributions in PyMC are specified. As an example, fields like psychology and astrophysics have complex likelihood functions for particular processes that may require numerical approximation.
Implementing the beta variable above as a Continuous subclass is shown below, along with an associated RandomVariable object, an instance of which becomes an attribute of the distribution.
class BetaRV(at.random.op.RandomVariable):
name = "beta"
ndim_supp = 0
ndims_params = []
dtype = "floatX"
@classmethod
def rng_fn(cls, rng, size):
raise NotImplementedError("Cannot sample from beta variable")
beta = BetaRV()
class Beta(pm.Continuous):
rv_op = beta
@classmethod
def dist(cls, mu=0, **kwargs):
mu = at.as_tensor_variable(mu)
return super().dist([mu], **kwargs)
def logp(self, value):
mu = self.mu
return beta_logp(value - mu)
def beta_logp(value):
return -1.5 * at.log(1 + (value) ** 2)
with pm.Model() as model:
beta = Beta("beta", mu=0)
If your logp can not be expressed in PyTensor, you can decorate the function with as_op as follows: @as_op(itypes=[at.dscalar], otypes=[at.dscalar]). Note, that this will create a blackbox Python function that will be much slower and not provide the gradients necessary for e.g. NUTS.
Discussion#
Probabilistic programming is an emerging paradigm in statistical learning, of which Bayesian modeling is an important sub-discipline. The signature characteristics of probabilistic programming–specifying variables as probability distributions and conditioning variables on other variables and on observations–makes it a powerful tool for building models in a variety of settings, and over a range of model complexity. Accompanying the rise of probabilistic programming has been a burst of innovation in fitting methods for Bayesian models that represent notable improvement over existing MCMC methods. Yet, despite this expansion, there are few software packages available that have kept pace with the methodological innovation, and still fewer that allow non-expert users to implement models.
PyMC provides a probabilistic programming platform for quantitative researchers to implement statistical models flexibly and succinctly. A large library of statistical distributions and several pre-defined fitting algorithms allows users to focus on the scientific problem at hand, rather than the implementation details of Bayesian modeling. The choice of Python as a development language, rather than a domain-specific language, means that PyMC users are able to work interactively to build models, introspect model objects, and debug or profile their work, using a dynamic, high-level programming language that is easy to learn. The modular, object-oriented design of PyMC means that adding new fitting algorithms or other features is straightforward. In addition, PyMC comes with several features not found in most other packages, most notably Hamiltonian-based samplers as well as automatic transforms of constrained random variables which is only offered by Stan. Unlike Stan, however, PyMC supports discrete variables as well as non-gradient based sampling algorithms like Metropolis-Hastings and Slice sampling.
Development of PyMC is an ongoing effort and several features are planned for future versions. Most notably, variational inference techniques are often more efficient than MCMC sampling, at the cost of generalizability. More recently, however, black-box variational inference algorithms have been developed, such as automatic differentiation variational inference (ADVI; Kucukelbir et al., 2017). This algorithm is slated for addition to PyMC. As an open-source scientific computing toolkit, we encourage researchers developing new fitting algorithms for Bayesian models to provide reference implementations in PyMC. Since samplers can be written in pure Python code, they can be implemented generally to make them work on arbitrary PyMC models, giving authors a larger audience to put their methods into use.
References#
Bastien, F., Lamblin, P., Pascanu, R., Bergstra, J., Goodfellow, I., Bergeron, A., Bouchard, N., Warde-Farley, D., and Bengio, Y. (2012) “Theano: new features and speed improvements”. NIPS 2012 deep learning workshop.
Bergstra, J., Breuleux, O., Bastien, F., Lamblin, P., Pascanu, R., Desjardins, G., Turian, J., Warde-Farley, D., and Bengio, Y. (2010) “Theano: A CPU and GPU Math Expression Compiler”. Proceedings of the Python for Scientific Computing Conference (SciPy) 2010. June 30 - July 3, Austin, TX
Duane, S., Kennedy, A. D., Pendleton, B. J., and Roweth, D. (1987) “Hybrid Monte Carlo”, Physics Letters, vol. 195, pp. 216-222.
Hoffman, M. D., & Gelman, A. (2014). The No-U-Turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. The Journal of Machine Learning Research, 30.
Jarrett, R.G. A note on the intervals between coal mining disasters. Biometrika, 66:191–193, 1979.
Lunn, D.J., Thomas, A., Best, N., and Spiegelhalter, D. (2000) WinBUGS – a Bayesian modelling framework: concepts, structure, and extensibility. Statistics and Computing, 10:325–337.
Neal, R.M. Slice sampling. Annals of Statistics. (2003). doi:10.2307/3448413. Patil, A., D. Huard and C.J. Fonnesbeck. (2010) PyMC: Bayesian Stochastic Modelling in Python. Journal of Statistical Software, 35(4), pp. 1-81
Stan Development Team. (2014). Stan: A C++ Library for Probability and Sampling, Version 2.5.0.
%load_ext watermark
%watermark -n -u -v -iv -w -p xarray
Last updated: Mon Apr 15 2024
Python implementation: CPython
Python version : 3.11.8
IPython version : 8.22.2
xarray: 2024.3.0
pytensor : 2.20.0
pymc : 5.13.1+0.g34c2d31.dirty
numpy : 1.26.4
pandas : 2.2.2
matplotlib: 3.8.4
arviz : 0.18.0
Watermark: 2.4.3